Genomic insights of Pannonibacter phragmitetus strain 31801 isolated from a patient with a liver abscess
- PMID: 28857514
- PMCID: PMC5727363
- DOI: 10.1002/mbo3.515
Genomic insights of Pannonibacter phragmitetus strain 31801 isolated from a patient with a liver abscess
Abstract
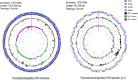
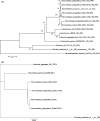
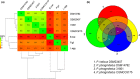
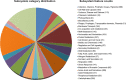
Pannonibacter phragmitetus is a bioremediation reagent for the detoxification of heavy metals and polycyclic aromatic compounds (PAHs) while it rarely infects healthy populations. However, infection by the opportunistic pathogen P. phragmitetus complicates diagnosis and treatments, and poses a serious threat to immunocompromised patients owing to its multidrug resistance. Unfortunately, genome features, antimicrobial resistance, and virulence potentials in P. phragmitetus have not been reported before. A predominant colony (31801) was isolated from a liver abscess patient, indicating that it accounted for the infection. To investigate its infection mechanism(s) in depth, we sequenced this bacterial genome and tested its antimicrobial resistance. Average nucleotide identity (ANI) analysis assigned the bacterium to the species P. phragmitetus (ANI, >95%). Comparative genomics analyses among Pannonibacter spp. representing the different living niches were used to describe the Pannonibacter pan-genomes and to examine virulence factors, prophages, CRISPR arrays, and genomic islands. Pannonibacter phragmitetus 31801 consisted of one chromosome and one plasmid, while the plasmid was absent in other Pannonibacter isolates. Pannonibacter phragmitetus 31801 may have a great infection potential because a lot of genes encoding toxins, flagellum formation, iron uptake, and virulence factor secretion systems in its genome. Moreover, the genome has 24 genomic islands and 2 prophages. A combination of antimicrobial susceptibility tests and the detailed antibiotic resistance gene analysis provide useful information about the drug resistance mechanisms and therefore can be used to guide the treatment strategy for the bacterial infection.
Keywords: Pannonibacter phragmitetus; antibiotic resistance mechanism; comparative genomic analysis; genomic sequencing; pathogenesis.
© 2017 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures

Similar articles
-
Analysis of resistance genes of clinical Pannonibacter phragmitetus strain 31801 by complete genome sequencing.Arch Microbiol. 2018 Sep;200(7):1101-1109. doi: 10.1007/s00203-018-1522-2. Epub 2018 May 14. Arch Microbiol. 2018. PMID: 29761214
-
Pannonibacter carbonis sp. nov., isolated from coal mine water.Int J Syst Evol Microbiol. 2018 Jun;68(6):2042-2047. doi: 10.1099/ijsem.0.002794. Epub 2018 Apr 25. Int J Syst Evol Microbiol. 2018. PMID: 29693544
-
Pannonibacter indica sp. nov., a highly arsenate-tolerant bacterium isolated from a hot spring in India.Arch Microbiol. 2013 Jan;195(1):1-8. doi: 10.1007/s00203-012-0840-z. Epub 2012 Sep 1. Arch Microbiol. 2013. PMID: 22940883
-
Liver Abscess Caused by Pannonibacter phragmitetus: Case Report and Literature Review.Front Med (Lausanne). 2017 Apr 25;4:48. doi: 10.3389/fmed.2017.00048. eCollection 2017. Front Med (Lausanne). 2017. PMID: 28487855 Free PMC article.
-
Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms.Front Cell Infect Microbiol. 2017 Nov 21;7:483. doi: 10.3389/fcimb.2017.00483. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29209595 Free PMC article. Review.
Cited by
-
Identification and denitrification characteristics of a salt-tolerant denitrifying bacterium Pannonibacter phragmitetus F1.AMB Express. 2019 Dec 3;9(1):193. doi: 10.1186/s13568-019-0918-y. AMB Express. 2019. PMID: 31797109 Free PMC article.
-
Genomic characteristics of antimicrobial resistance and virulence factors of carbapenem-resistant Stutzerimonas nitrititolerans isolated from the clinical specimen.BMC Microbiol. 2024 Oct 3;24(1):386. doi: 10.1186/s12866-024-03546-4. BMC Microbiol. 2024. PMID: 39358682 Free PMC article.
-
Hemodialysis catheter-related infection caused by Pannonibacter phragmitetus: a rare case report in China.Front Cell Infect Microbiol. 2022 Jul 22;12:926154. doi: 10.3389/fcimb.2022.926154. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35959368 Free PMC article.
References
-
- Bandyopadhyay, S. , Schumann, P. , & Das, S. K. (2013). Pannonibacter indica sp. nov., a highly arsenate‐tolerant bacterium isolated from a hot spring in India. Archives of Microbiology, 195, 1–8. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

