X Chromosome Evolution in Cetartiodactyla
- PMID: 28858207
- PMCID: PMC5615350
- DOI: 10.3390/genes8090216
X Chromosome Evolution in Cetartiodactyla
Abstract
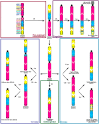
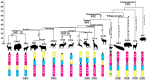
The phenomenon of a remarkable conservation of the X chromosome in eutherian mammals has been first described by Susumu Ohno in 1964. A notable exception is the cetartiodactyl X chromosome, which varies widely in morphology and G-banding pattern between species. It is hypothesized that this sex chromosome has undergone multiple rearrangements that changed the centromere position and the order of syntenic segments over the last 80 million years of Cetartiodactyla speciation. To investigate its evolution we have selected 26 evolutionarily conserved bacterial artificial chromosome (BAC) clones from the cattle CHORI-240 library evenly distributed along the cattle X chromosome. High-resolution BAC maps of the X chromosome on a representative range of cetartiodactyl species from different branches: pig (Suidae), alpaca (Camelidae), gray whale (Cetacea), hippopotamus (Hippopotamidae), Java mouse-deer (Tragulidae), pronghorn (Antilocapridae), Siberian musk deer (Moschidae), and giraffe (Giraffidae) were obtained by fluorescent in situ hybridization. To trace the X chromosome evolution during fast radiation in specious families, we performed mapping in several cervids (moose, Siberian roe deer, fallow deer, and Pere David's deer) and bovid (muskox, goat, sheep, sable antelope, and cattle) species. We have identified three major conserved synteny blocks and rearrangements in different cetartiodactyl lineages and found that the recently described phenomenon of the evolutionary new centromere emergence has taken place in the X chromosome evolution of Cetartiodactyla at least five times. We propose the structure of the putative ancestral cetartiodactyl X chromosome by reconstructing the order of syntenic segments and centromere position for key groups.
Keywords: Pecora; Ruminantia; cattle bacterial artificial chromosome (BAC) clones; centromere reposition; fluorescent in situ hybridization (FISH); intrachromosomal rearrangements; inversion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Murphy W.J., Larkin D.M., Everts-Van Der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. - DOI - PubMed
-
- Graphodatsky A.S. Conserved and variable elements of mammalian chromosomes. Cytogenet. Anim. 1989:95–124.
LinkOut - more resources
Full Text Sources
Other Literature Sources

