Flavin monooxygenases regulate Caenorhabditis elegans axon guidance and growth cone protrusion with UNC-6/Netrin signaling and Rac GTPases
- PMID: 28859089
- PMCID: PMC5597259
- DOI: 10.1371/journal.pgen.1006998
Flavin monooxygenases regulate Caenorhabditis elegans axon guidance and growth cone protrusion with UNC-6/Netrin signaling and Rac GTPases
Abstract
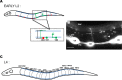
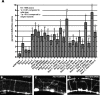
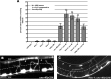
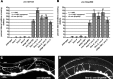
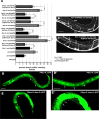
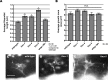
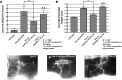
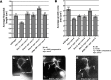
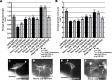
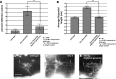
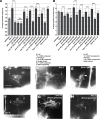
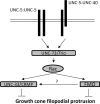
The guidance cue UNC-6/Netrin regulates both attractive and repulsive axon guidance. Our previous work showed that in C. elegans, the attractive UNC-6/Netrin receptor UNC-40/DCC stimulates growth cone protrusion, and that the repulsive receptor, an UNC-5:UNC-40 heterodimer, inhibits growth cone protrusion. We have also shown that inhibition of growth cone protrusion downstream of the UNC-5:UNC-40 repulsive receptor involves Rac GTPases, the Rac GTP exchange factor UNC-73/Trio, and the cytoskeletal regulator UNC-33/CRMP, which mediates Semaphorin-induced growth cone collapse in other systems. The multidomain flavoprotein monooxygenase (FMO) MICAL (Molecule Interacting with CasL) also mediates growth cone collapse in response to Semaphorin by directly oxidizing F-actin, resulting in depolymerization. The C. elegans genome does not encode a multidomain MICAL-like molecule, but does encode five flavin monooxygenases (FMO-1, -2, -3, -4, and 5) and another molecule, EHBP-1, similar to the non-FMO portion of MICAL. Here we show that FMO-1, FMO-4, FMO-5, and EHBP-1 may play a role in UNC-6/Netrin directed repulsive guidance mediated through UNC-40 and UNC-5 receptors. Mutations in fmo-1, fmo-4, fmo-5, and ehbp-1 showed VD/DD axon guidance and branching defects, and variably enhanced unc-40 and unc-5 VD/DD axon guidance defects. Developing growth cones in vivo of fmo-1, fmo-4, fmo-5, and ehbp-1 mutants displayed excessive filopodial protrusion, and transgenic expression of FMO-5 inhibited growth cone protrusion. Mutations suppressed growth cone inhibition caused by activated UNC-40 and UNC-5 signaling, and activated Rac GTPase CED-10 and MIG-2, suggesting that these molecules are required downstream of UNC-6/Netrin receptors and Rac GTPases. From these studies we conclude that FMO-1, FMO-4, FMO-5, and EHBP-1 represent new players downstream of UNC-6/Netrin receptors and Rac GTPases that inhibit growth cone filopodial protrusion in repulsive axon guidance.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Pak CW, Flynn KC, Bamburg JR. Actin-binding proteins take the reins in growth cones. Nat Rev Neurosci. 2008;9(2):136–47. doi: 10.1038/nrn2236 . - DOI - PubMed
-
- Gallo G, Letourneau PC. Regulation of growth cone actin filaments by guidance cues. J Neurobiol. 2004;58(1):92–102. doi: 10.1002/neu.10282 . - DOI - PubMed
-
- Zhou FQ, Cohan CS. How actin filaments and microtubules steer growth cones to their targets. J Neurobiol. 2004;58(1):84–91. doi: 10.1002/neu.10278 . - DOI - PubMed
-
- Omotade OF, Pollitt SL, Zheng JQ. Actin-based growth cone motility and guidance. Mol Cell Neurosci. 2017. doi: 10.1016/j.mcn.2017.03.001 . - DOI - PMC - PubMed
-
- Mortimer D, Fothergill T, Pujic Z, Richards LJ, Goodhill GJ. Growth cone chemotaxis. Trends Neurosci. 2008. doi: 10.1016/j.tins.2007.11.008 . - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

