MYT1L mutations cause intellectual disability and variable obesity by dysregulating gene expression and development of the neuroendocrine hypothalamus
- PMID: 28859103
- PMCID: PMC5597252
- DOI: 10.1371/journal.pgen.1006957
MYT1L mutations cause intellectual disability and variable obesity by dysregulating gene expression and development of the neuroendocrine hypothalamus
Abstract
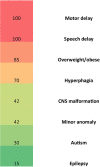
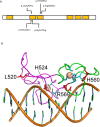
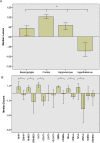
Deletions at chromosome 2p25.3 are associated with a syndrome consisting of intellectual disability and obesity. The smallest region of overlap for deletions at 2p25.3 contains PXDN and MYT1L. MYT1L is expressed only within the brain in humans. We hypothesized that single nucleotide variants (SNVs) in MYT1L would cause a phenotype resembling deletion at 2p25.3. To examine this we sought MYT1L SNVs in exome sequencing data from 4, 296 parent-child trios. Further variants were identified through a genematcher-facilitated collaboration. We report 9 patients with MYT1L SNVs (4 loss of function and 5 missense). The phenotype of SNV carriers overlapped with that of 2p25.3 deletion carriers. To identify the transcriptomic consequences of MYT1L loss of function we used CRISPR-Cas9 to create a knockout cell line. Gene Ontology analysis in knockout cells demonstrated altered expression of genes that regulate gene expression and that are localized to the nucleus. These differentially expressed genes were enriched for OMIM disease ontology terms "mental retardation". To study the developmental effects of MYT1L loss of function we created a zebrafish knockdown using morpholinos. Knockdown zebrafish manifested loss of oxytocin expression in the preoptic neuroendocrine area. This study demonstrates that MYT1L variants are associated with syndromic obesity in humans. The mechanism is related to dysregulated expression of neurodevelopmental genes and altered development of the neuroendocrine hypothalamus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Van der Klaauw AA, Farooqi IS. The hunger genes: pathways to obesity. Cell. 2015;161(1): 119–132. doi: 10.1016/j.cell.2015.03.008 - DOI - PubMed
-
- Rosenfeld JA, Patel A. Chromosomal microarrays: understanding genetics of neurodevelopmental disorders and congenital anomalies. J Pediatr Genet. 2017; 6 (1): 42–50. doi: 10.1055/s-0036-1584306 - DOI - PMC - PubMed
-
- Wright CF, Fitzgerald TW, Jones WD, Clayton S, McRae JF, Kogelenberg MV, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015; 385 (9975): 1305–1314. doi: 10.1016/S0140-6736(14)61705-0 - DOI - PMC - PubMed
-
- Kaur Y, de Souza RJ, Gibson WT, Meyre D. A systematic review of genetic syndromes with obesity. Obes Rev. 2017; doi: 10.1111/obr.12531 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

