Transcript profiling of genes expressed during fibre development in diploid cotton (Gossypium arboreum L.)
- PMID: 28859611
- PMCID: PMC5580217
- DOI: 10.1186/s12864-017-4066-y
Transcript profiling of genes expressed during fibre development in diploid cotton (Gossypium arboreum L.)
Abstract
Background: Cotton fibre is a single cell and it is one of the best platforms for unraveling the genes express during various stages of fibre development. There are reports devoted to comparative transcriptome study on fiber cell initiation and elongation in tetraploid cultivated cotton. However, in the present investigation, comparative transcriptome study was made in diploid cultivated cotton using isogenic fuzzy-lintless (Fl) and normal fuzzy linted (FL) lines belong to Gossypium arboreum, diploid species at two stages, 0 and 10 dpa (days post anthesis), using Affymetrix cotton GeneChip genome array.
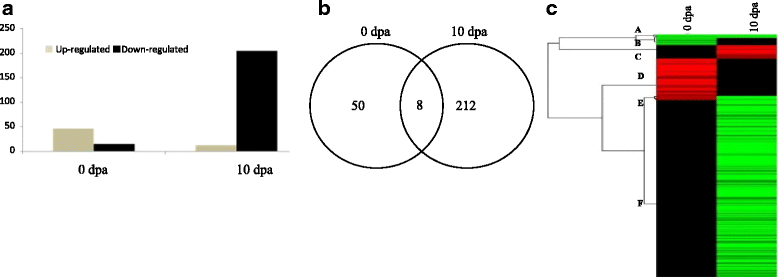
Result: Scanning electron microscopy (SEM) analysis uncovered the occurrence of few fibre cell initials in the Fl line as compared to many in Normal FL at -2 and 0 dpa. However, at 10 dpa there were no fibre cells found elongated in Fl but many elongated cells were found in FL line. Up-regulation of transcription factors, AP2-EREBP, C2H2, C3H, HB and WRKY was observed at 0 dpa whereas in 10 dpa transcription factors, AP2-EREBP, AUX/IAA, bHLH, C2H2, C3H, HB, MYB, NAC, Orphans, PLATZ and WRKY were found down regulated in Fl line. These transcription factors were mainly involved in metabolic pathways such as phytohormone signaling, energy metabolism of cell, fatty acid metabolism, secondary metabolism and other signaling pathways and are related directly or indirectly in fiber development. Quantitative real-time PCR was performed to check fold up or down-regulation of these genes and transcription factors (TFs) down regulated in mutants as compared to normal at 0 and 10 dpa.
Conclusion: This study elucidates that the up-regulation of transcription factors like AP2-EREBP, C2H2, C3H, HB, WRKY and phytohormone signaling genes at 0 dpa and their down-regulation at the 10 dpa might have constrain the fibre elongation in fuzzy-lintless line. Along with this the down-regulation of genes involved in synthesis of VLCFA chain, transcripts necessary for energy and cell wall metabolism, EXPANSINs, arabinogalactan proteins (AGPs), tubulin might also be the probable reason for reduced growth of fibres in the Fl. Plant receptor-like kinases (RLKs), Leucine Rich Repeats) LRR- family protein and signal transduction coding for mitogen-activated protein kinase (MAPK) cascade, have been engaged in coordination of cell elongation and SCW biosynthesis, down-regulation of these might loss the function leads to reduced fibre growth.
Keywords: Diploid cotton; Microarray; Transcription factors; qRT PCR.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




Similar articles
-
Functional genomics of fuzzless-lintless mutant of Gossypium hirsutum L. cv. MCU5 reveal key genes and pathways involved in cotton fibre initiation and elongation.BMC Genomics. 2012 Nov 14;13:624. doi: 10.1186/1471-2164-13-624. BMC Genomics. 2012. PMID: 23151214 Free PMC article.
-
Gene expression profile analysis of Ligon lintless-1 (Li1) mutant reveals important genes and pathways in cotton leaf and fiber development.Gene. 2014 Feb 10;535(2):273-85. doi: 10.1016/j.gene.2013.11.017. Epub 2013 Nov 23. Gene. 2014. PMID: 24279997
-
Genetic Identification and Transcriptome Analysis of Lintless and Fuzzless Traits in Gossypium arboreum L.Int J Mol Sci. 2020 Feb 29;21(5):1675. doi: 10.3390/ijms21051675. Int J Mol Sci. 2020. PMID: 32121400 Free PMC article.
-
Gene expression changes and early events in cotton fibre development.Ann Bot. 2007 Dec;100(7):1391-401. doi: 10.1093/aob/mcm232. Epub 2007 Sep 27. Ann Bot. 2007. PMID: 17905721 Free PMC article. Review.
-
A comprehensive overview of cotton genomics, biotechnology and molecular biological studies.Sci China Life Sci. 2023 Oct;66(10):2214-2256. doi: 10.1007/s11427-022-2278-0. Epub 2023 Mar 6. Sci China Life Sci. 2023. PMID: 36899210 Review.
Cited by
-
Identification of Introgressed Alleles Conferring High Fiber Quality Derived From Gossypium barbadense L. in Secondary Mapping Populations of G. hirsutum L.Front Plant Sci. 2018 Jul 18;9:1023. doi: 10.3389/fpls.2018.01023. eCollection 2018. Front Plant Sci. 2018. PMID: 30073008 Free PMC article.
-
Artificial Biopolymers Derived from Transgenic Plants: Applications and Properties-A Review.Int J Mol Sci. 2024 Dec 19;25(24):13628. doi: 10.3390/ijms252413628. Int J Mol Sci. 2024. PMID: 39769390 Free PMC article. Review.
-
Genome-Wide Identification and Expression Analysis of the PLATZ Transcription Factor in Tomato.Plants (Basel). 2023 Jul 13;12(14):2632. doi: 10.3390/plants12142632. Plants (Basel). 2023. PMID: 37514247 Free PMC article.
-
Arginine Decarboxylase Gene ADC2 Regulates Fiber Elongation in Cotton.Genes (Basel). 2022 Apr 28;13(5):784. doi: 10.3390/genes13050784. Genes (Basel). 2022. PMID: 35627169 Free PMC article.
-
Genome-Wide Identification and Salt Stress-Responsive Expression Analysis of the GmPLATZ Gene Family in Soybean (Glycine max L.).Plants (Basel). 2025 Jun 30;14(13):2004. doi: 10.3390/plants14132004. Plants (Basel). 2025. PMID: 40648010 Free PMC article.
References
-
- Farooq J, Farooq A, Rizwan M, Petrescu-Mag IV, Ali MA, Mahmood K, Batool A. Cotton fibers: attributes of specialized cells and factors affecting them. AES Bioflux. 2015;7(3):369–382.
-
- Wendel JF, Brubaker CL, Percival AE. Genetic diversity in Gossypium hirsutum and the origin of upland cotton. Am J Bot. 1992;79:1291–1310. doi: 10.2307/2445058. - DOI
-
- Fan L, Hu W, Yang Y, Li B. Plant Special Cell - Cotton Fiber. Dr. John Mworia editors. Botany. InTech, Janeza Trdine 9, 51000 Rijeka, Croatia: 2012. p. 211–226.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

