A streamlined method for analysing genome-wide DNA methylation patterns from low amounts of FFPE DNA
- PMID: 28859641
- PMCID: PMC5580311
- DOI: 10.1186/s12920-017-0290-1
A streamlined method for analysing genome-wide DNA methylation patterns from low amounts of FFPE DNA
Abstract
Background: Formalin fixed paraffin embedded (FFPE) tumor samples are a major source of DNA from patients in cancer research. However, FFPE is a challenging material to work with due to macromolecular fragmentation and nucleic acid crosslinking. FFPE tissue particularly possesses challenges for methylation analysis and for preparing sequencing-based libraries relying on bisulfite conversion. Successful bisulfite conversion is a key requirement for sequencing-based methylation analysis.
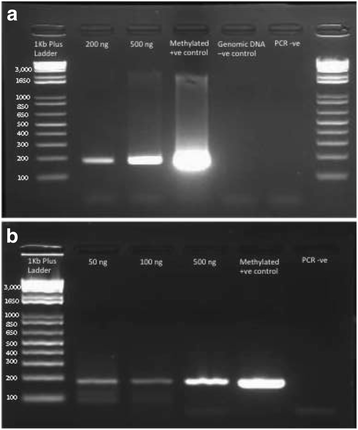
Methods: Here we describe a complete and streamlined workflow for preparing next generation sequencing libraries for methylation analysis from FFPE tissues. This includes, counting cells from FFPE blocks and extracting DNA from FFPE slides, testing bisulfite conversion efficiency with a polymerase chain reaction (PCR) based test, preparing reduced representation bisulfite sequencing libraries and massively parallel sequencing.
Results: The main features and advantages of this protocol are: An optimized method for extracting good quality DNA from FFPE tissues. An efficient bisulfite conversion and next generation sequencing library preparation protocol that uses 50 ng DNA from FFPE tissue. Incorporation of a PCR-based test to assess bisulfite conversion efficiency prior to sequencing.
Conclusions: We provide a complete workflow and an integrated protocol for performing DNA methylation analysis at the genome-scale and we believe this will facilitate clinical epigenetic research that involves the use of FFPE tissue.
Keywords: Bisulfite sequencing; DNA extraction; DNA methylation; FFPE tissue; Genome-wide; RRBS.
Conflict of interest statement
Ethics approval and consent to participate
Collection of tissue samples and consent of patients were performed according to the protocol and approval from Health and Disability Ethics Committee (Ethics protocol number: LRS1102002).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Chatterjee A, Rodger EJ, Eccles MR. Epigenetic drivers of tumourigenesis and cancer metastasis. Semin Cancer Biol. 2017. doi:10.1016/j.semcancer.2017.08.004. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

