The respiratory tract microbial biogeography in alcohol use disorder
- PMID: 28860145
- PMCID: PMC5866426
- DOI: 10.1152/ajplung.00277.2017
The respiratory tract microbial biogeography in alcohol use disorder
Abstract
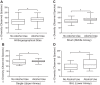
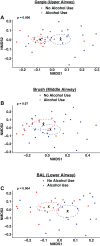
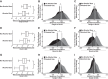
Individuals with alcohol use disorders (AUDs) are at an increased risk of pneumonia and acute respiratory distress syndrome. Data of the lung microbiome in the setting of AUDs are lacking. The objective of this study was to determine the microbial biogeography of the upper and lower respiratory tract in individuals with AUDs compared with non-AUD subjects. Gargle, protected bronchial brush, and bronchoalveolar lavage specimens were collected during research bronchoscopies. Bacterial 16S gene sequencing and phylogenetic analysis was performed, and the alterations to the respiratory tract microbiota and changes in microbial biogeography were determined. The microbial structure of the upper and lower respiratory tract was significantly altered in subjects with AUDs compared with controls. Subjects with AUD have greater microbial diversity [ P < 0.0001, effect size = 16 ± 1.7 observed taxa] and changes in microbial species relative abundances. Furthermore, microbial communities in the upper and lower respiratory tract displayed greater similarity in subjects with AUDs. Alcohol use is associated with an altered composition of the respiratory tract microbiota. Subjects with AUDs demonstrate convergence of the microbial phylogeny and taxonomic communities between distinct biogeographical sites within the respiratory tract. These results support a mechanistic pathway potentially explaining the increased incidence of pneumonia and lung diseases in patients with AUDs.
Keywords: alcohol; biogeography; lung; microbiome; respiratory.
Figures




References
-
- Beck JM, Schloss PD, Venkataraman A, Twigg H III, Jablonski KA, Bushman FD, Campbell TB, Charlson ES, Collman RG, Crothers K, Curtis JL, Drews KL, Flores SC, Fontenot AP, Foulkes MA, Frank I, Ghedin E, Huang L, Lynch SV, Morris A, Palmer BE, Schmidt TM, Sodergren E, Weinstock GM, Young VB; Lung HIV Microbiome Project . Multicenter comparison of lung and oral microbiomes of HIV-infected and HIV-uninfected individuals. Am J Respir Crit Care Med 192: 1335–1344, 2015. doi: 10.1164/rccm.201501-0128OC. - DOI - PMC - PubMed
-
- Berkowitz H, Reichel J, Shim C. The effect of ethanol on the cough reflex. Clin Sci Mol Med 45: 527–531, 1973. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

