Multiple miRNAs jointly regulate the biosynthesis of ecdysteroid in the holometabolous insects, Chilo suppressalis
- PMID: 28860304
- PMCID: PMC5689003
- DOI: 10.1261/rna.061408.117
Multiple miRNAs jointly regulate the biosynthesis of ecdysteroid in the holometabolous insects, Chilo suppressalis
Abstract
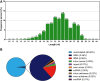
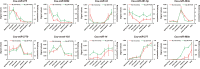
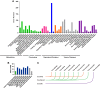
The accurate rise and fall of active hormones is important for insect development. The ecdysteroids must be cleared in a timely manner. However, the mechanism of suppressing the ecdysteroid biosynthesis at the right time remains unclear. Here, we sequenced a small RNA library of Chilo suppressalis and identified 300 miRNAs in this notorious rice insect pest. Microarray analysis yielded 54 differentially expressed miRNAs during metamorphosis development. Target prediction and in vitro dual-luciferase assays confirmed that seven miRNAs (two conserved and five novel miRNAs) jointly targeted three Halloween genes in the ecdysteroid biosynthesis pathway. Overexpression of these seven miRNAs reduced the titer of 20-hydroxyecdysone (20E), induced mortality, and retarded development, which could be rescued by treatment with 20E. Comparative analysis indicated that the miRNA regulation of metamorphosis development is a conserved process but that the miRNAs involved are highly divergent. In all, we present evidence that both conserved and lineage-specific miRNAs have crucial roles in regulating development in insects by controlling ecdysteroid biosynthesis, which is important for ensuring developmental convergence and evolutionary diversity.
Keywords: development; ecdysteroid; insect; metamorphosis; miRNA.
© 2017 He et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures







References
-
- Asgari S. 2013. MicroRNA functions in insects. Insect Biochem Mol Biol 43: 388–397. - PubMed
-
- Belles X. 2017. MicroRNAs and the evolution of insect metamorphosis. Annu Rev Entomol 62: 111–125. - PubMed
-
- Belles X, Cristino AS, Tanaka ED, Rubio M, Piulachs M-D. 2011. Insect microRNAs: from molecular mechanisms to biological roles. In Insect molecular biology and biochemistry (ed. Gilbert LI), pp. 30–56. ElsevierAcademic Press, Amsterdam, Netherlands.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
