Inert Gas Deactivates Protein Activity by Aggregation
- PMID: 28860621
- PMCID: PMC5579012
- DOI: 10.1038/s41598-017-10678-3
Inert Gas Deactivates Protein Activity by Aggregation
Abstract
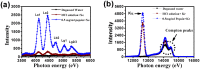
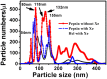
Biologically inert gases play important roles in the biological functionality of proteins. However, researchers lack a full understanding of the effects of these gases since they are very chemically stable only weakly absorbed by biological tissues. By combining X-ray fluorescence, particle sizing and molecular dynamics (MD) simulations, this work shows that the aggregation of these inert gases near the hydrophobic active cavity of pepsin should lead to protein deactivation. Micro X-ray fluorescence spectra show that a pepsin solution can contain a high concentration of Xe or Kr after gassing, and that the gas concentrations decrease quickly with degassing time. Biological activity experiments indicate a reversible deactivation of the protein during this gassing and degassing. Meanwhile, the nanoparticle size measurements reveal a higher number of "nanoparticles" in gas-containing pepsin solution, also supporting the possible interaction between inert gases and the protein. Further, MD simulations indicate that gas molecules can aggregate into a tiny bubble shape near the hydrophobic active cavity of pepsin, suggesting a mechanism for reducing their biological function.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

