Structural and Functional View of Polypharmacology
- PMID: 28860623
- PMCID: PMC5579063
- DOI: 10.1038/s41598-017-10012-x
Structural and Functional View of Polypharmacology
Abstract
Protein domains mediate drug-protein interactions and this principle can guide the design of multi-target drugs i.e. polypharmacology. In this study, we associate multi-target drugs with CATH functional families through the overrepresentation of targets of those drugs in CATH functional families. Thus, we identify CATH functional families that are currently enriched in drugs (druggable CATH functional families) and we use the network properties of these druggable protein families to analyse their association with drug side effects. Analysis of selected druggable CATH functional families, enriched in drug targets, show that relatives exhibit highly conserved drug binding sites. Furthermore, relatives within druggable CATH functional families occupy central positions in a human protein functional network, cluster together forming network neighbourhoods and are less likely to be within proteins associated with drug side effects. Our results demonstrate that CATH functional families can be used to identify drug-target interactions, opening a new research direction in target identification.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
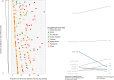
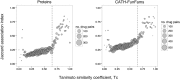
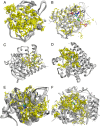
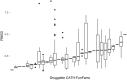
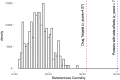
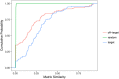
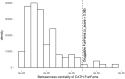
References
-
- Anighoro, A., Bajorath, J. & Rastelli, G. Polypharmacology: Challenges and Opportunities in Drug Discovery. J Med Chem, doi:10.1021/jm5006463 (2014). - PubMed
-
- Moya-García AA, Morilla I, Ranea JAG. Oncogenic Signalling Networks and Polypharmacology as Paradigms to Cope with Cancer Heterogeneity. Current Proteomics. 2014;11:1–8. doi: 10.2174/157016461101140709105405. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

