Small, Enigmatic Plasmids of the Nosocomial Pathogen, Acinetobacter baumannii: Good, Bad, Who Knows?
- PMID: 28861061
- PMCID: PMC5559437
- DOI: 10.3389/fmicb.2017.01547
Small, Enigmatic Plasmids of the Nosocomial Pathogen, Acinetobacter baumannii: Good, Bad, Who Knows?
Abstract
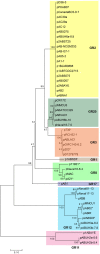
Acinetobacter baumannii is a Gram-negative nosocomial pathogen that has become a serious healthcare concern within a span of two decades due to its ability to rapidly acquire resistance to all classes of antimicrobial compounds. One of the key features of the A. baumannii genome is an open pan genome with a plethora of plasmids, transposons, integrons, and genomic islands, all of which play important roles in the evolution and success of this clinical pathogen, particularly in the acquisition of multidrug resistance determinants. An interesting genetic feature seen in majority of A. baumannii genomes analyzed is the presence of small plasmids that usually ranged from 2 to 10 kb in size, some of which harbor antibiotic resistance genes and homologs of plasmid mobilization genes. These plasmids are often overlooked when compared to their larger, conjugative counterparts that harbor multiple antibiotic resistance genes and transposable elements. In this mini-review, we will examine our current knowledge of these small A. baumannii plasmids and look into their genetic diversity and phylogenetic relationships. Some of these plasmids, such as the Rep-3 superfamily group and the pRAY-type, which has no recognizable replicase genes, are quite widespread among diverse A. baumannii clinical isolates worldwide, hinting at their usefulness to the lifestyle of this pathogen. Other small plasmids especially those from the Rep-1 superfamily are truly enigmatic, encoding only hypothetical proteins of unknown function, leading to the question of whether these small plasmids are "good" or "bad" to their host A. baumannii.
Keywords: Acinetobacter baumannii; Rep-1 superfamily; Rep-3 superfamily; antibiotic resistance genes; mobilizable plasmids; pRAY plasmids; small plasmids; toxin–antitoxin.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

