Unraveling the Fecal Microbiota and Metagenomic Functional Capacity Associated with Feed Efficiency in Pigs
- PMID: 28861066
- PMCID: PMC5559535
- DOI: 10.3389/fmicb.2017.01555
Unraveling the Fecal Microbiota and Metagenomic Functional Capacity Associated with Feed Efficiency in Pigs
Abstract
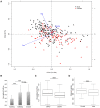
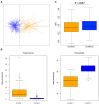
Gut microbiota plays fundamental roles in energy harvest, nutrient digestion, and intestinal health, especially in processing indigestible components of polysaccharides in diet. Unraveling the microbial taxa and functional capacity of gut microbiome associated with feed efficiency can provide important knowledge to improve pig feed efficiency in swine industry. In the current research, we studied the association of fecal microbiota with feed efficiency in 280 commercial Duroc pigs. All experimental pigs could be clustered into two enterotype-like groups. Different enterotypes showed the tendency of association with the feed efficiency (P = 0.07). We further identified 31 operational taxonomic units (OTUs) showing the potential associations with porcine feed efficiency. These OTUs were mainly annotated to the bacteria related to the metabolisms of dietary polysaccharides. Although we did not identify the RFI-associated bacterial species at FDR < 0.05 level, metagenomic sequencing analysis did find the distinct function capacities of gut microbiome between the high and low RFI pigs (FDR < 0.05). The KEGG orthologies related to nitrogen metabolism, amino acid metabolism, and transport system, and eight KEGG pathways including glycine, serine, and threonine metabolism were positively associated with porcine feed efficiency. We inferred that gut microbiota might improve porcine feed efficiency through promoting intestinal health by the SCFAs produced by fermenting dietary polysaccharides and improving the utilization of dietary protein. The present results provided important basic knowledge for improving porcine feed efficiency through modulating gut microbiome.
Keywords: 16S rRNA gene; feed efficiency; gut microbiota; metagenome; swine.
Figures


References
-
- Armstrong T. A., Spears J. W., Crenshaw T. D., Nielsen F. H. (2000). Boron supplementation of a semipurified diet for weanling pigs improves feed efficiency and bone strength characteristics and alters plasma lipid metabolites. J. Nutr. 130, 2575–2581. - PubMed
-
- Cao K. A. L., Déjean S., González I. (2012). Unravelling ‘Omics’ Data with the R Package Mixomics. 1ères Rencontres, R.Hal-00717497, version 1.
LinkOut - more resources
Full Text Sources
Other Literature Sources

