Identification of Genes under Positive Selection Reveals Differences in Evolutionary Adaptation between Brown-Algal Species
- PMID: 28861104
- PMCID: PMC5559719
- DOI: 10.3389/fpls.2017.01429
Identification of Genes under Positive Selection Reveals Differences in Evolutionary Adaptation between Brown-Algal Species
Abstract
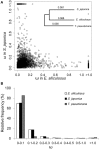
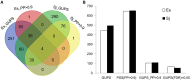
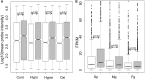
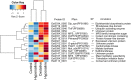
Brown algae are an important taxonomic group in coastal ecosystems. The model brown algal species Ectocarpus siliculosus and Saccharina japonica are closely related lineages. Despite their close phylogenetic relationship, they vary greatly in morphology and physiology. To obtain further insights into the evolutionary forces driving divergence in brown algae, we analyzed 3,909 orthologs from both species to identify Genes Under Positive Selection (GUPS). About 12% of the orthologs in each species were considered to be under positive selection. Many GUPS are involved in membrane transport, regulation of homeostasis, and sexual reproduction in the small sporophyte of E. siliculosus, which is known to have a complex life cycle and to occupy a wide range of habitats. Genes involved in photosynthesis and cell division dominated the group of GUPS in the large kelp of S. japonica, which might explain why this alga has evolved the ability to grow very rapidly and to form some of the largest sporophytes. A significant number of molecular chaperones (e.g., heat-shock proteins) involved in stress responses were identified to be under positive selection in both species, potentially indicating their important roles for macroalgae to cope with the relatively variable environment of coastal ecosystems. Moreover, analysis of previously published microarray data of E. siliculosus showed that many GUPS in E. siliculosus were responsive to stress conditions, such as oxidative and hyposaline stress, whereas our RNA-seq data of S. japonica showed that GUPS in this species were most highly expressed in large sporophytes, which supports the suggestion that selection largely acts on different sets of genes in both marcoalgal species, potentially reflecting their adaptation to different ecological niches.
Keywords: Ectocarpus siliculosus; Saccharina japonica; adaptive evolution; brown algae; positive selection.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

