Gene functioning and storage within a folded genome
- PMID: 28861108
- PMCID: PMC5575855
- DOI: 10.1186/s11658-017-0050-4
Gene functioning and storage within a folded genome
Abstract
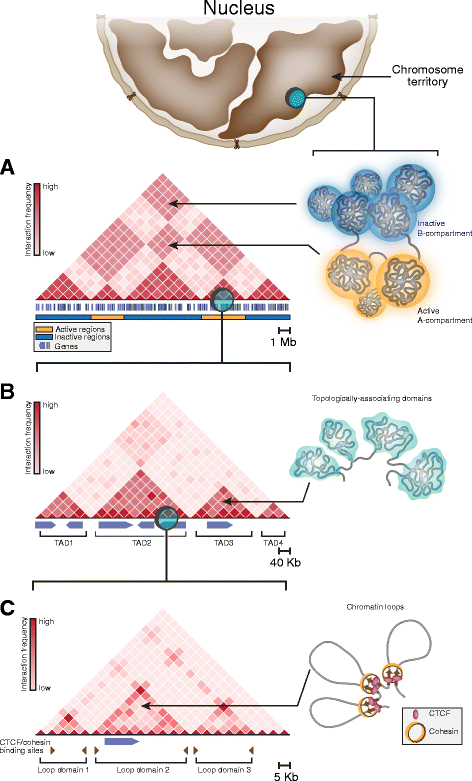
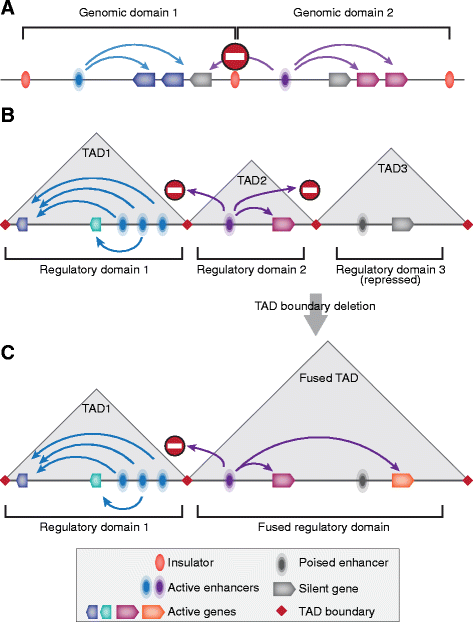
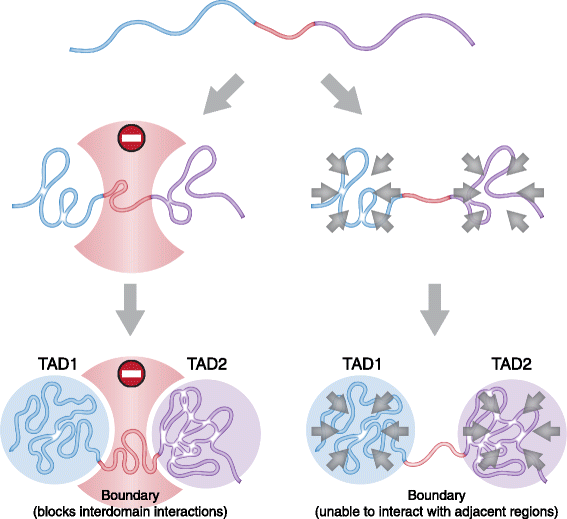
In mammals, genomic DNA that is roughly 2 m long is folded to fit the size of the cell nucleus that has a diameter of about 10 μm. The folding of genomic DNA is mediated via assembly of DNA-protein complex, chromatin. In addition to the reduction of genomic DNA linear dimensions, the assembly of chromatin allows to discriminate and to mark active (transcribed) and repressed (non-transcribed) genes. Consequently, epigenetic regulation of gene expression occurs at the level of DNA packaging in chromatin. Taking into account the increasing attention of scientific community toward epigenetic systems of gene regulation, it is very important to understand how DNA folding in chromatin is related to gene activity. For many years the hierarchical model of DNA folding was the most popular. It was assumed that nucleosome fiber (10-nm fiber) is folded into 30-nm fiber and further on into chromatin loops attached to a nuclear/chromosome scaffold. Recent studies have demonstrated that there is much less regularity in chromatin folding within the cell nucleus. The very existence of 30-nm chromatin fibers in living cells was questioned. On the other hand, it was found that chromosomes are partitioned into self-interacting spatial domains that restrict the area of enhancers action. Thus, TADs can be considered as structural-functional domains of the chromosomes. Here we discuss the modern view of DNA packaging within the cell nucleus in relation to the regulation of gene expression. Special attention is paid to the possible mechanisms of the chromatin fiber self-assembly into TADs. We discuss the model postulating that partitioning of the chromosome into TADs is determined by the distribution of active and inactive chromatin segments along the chromosome. This article was specially invited by the editors and represents work by leading researchers.
Keywords: Active chromatin; Enhancers; Epigenetic regulatory mechanisms; Hi-C; Self-organization; TADs.
Conflict of interest statement
Ethics approval and consent to participate
The authors state that no ethical approval was required for this work.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

