Genome-wide identification of histone H2A and histone variant H2A.Z-interacting proteins by bPPI-seq
- PMID: 28862252
- PMCID: PMC5630678
- DOI: 10.1038/cr.2017.112
Genome-wide identification of histone H2A and histone variant H2A.Z-interacting proteins by bPPI-seq
Abstract
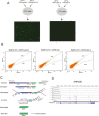
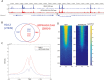
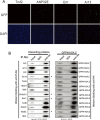
H2A is a nucleosome core subunit involved in organizing DNA into a chromatin structure that is often inaccessible to regulatory enzymes. Replacement of H2A by its variant H2A.Z renders chromatin accessible at enhancers and promoters. However, it remains unclear how H2A.Z functions so differently from canonical H2A. Here we report the genome-wide identification of proteins that directly interact with H2A and H2A.Z in vivo using a novel strategy, bPPI-seq. We show that bPPI-seq is a sensitive and robust technique to identify protein-protein interactions in vivo. Our data indicate that H2A.Z-interacting proteins and H2A-interacting proteins participate in distinct biological processes. In contrast to H2A-interacting proteins, the H2A.Z-interacting proteins are involved in transcriptional regulation. We found that the transcription factor Osr1 interacts with H2A.Z both in vitro and in vivo. It also mediates H2A.Z incorporation to a large number of target sites and regulates gene expression. Our data indicate that bPPI-seq can be widely applied to identify genome-wide interacting proteins under physiological conditions.
Figures




References
-
- Faast R, Thonglairoam V, Schulz TC, et al. Histone variant H2A.Z is required for early mammalian development. Curr Biol 2001; 11:1183–1187. - PubMed
-
- Clarkson MJ, Wells JR, Gibson F, Saint R, Tremethick DJ. Regions of variant histone His2AvD required for Drosophila development. Nature 1999; 399:694–697. - PubMed
-
- Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129:823–837. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

