Automated Tracking of Cell Migration with Rapid Data Analysis
- PMID: 28862338
- PMCID: PMC5654555
- DOI: 10.1002/cpcb.28
Automated Tracking of Cell Migration with Rapid Data Analysis
Abstract
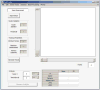
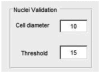
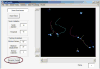
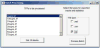
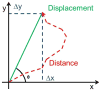
Cell migration is essential for many biological processes including development, wound healing, and metastasis. However, studying cell migration often requires the time-consuming and labor-intensive task of manually tracking cells. To accelerate the task of obtaining coordinate positions of migrating cells, we have developed a graphical user interface (GUI) capable of automating the tracking of fluorescently labeled nuclei. This GUI provides an intuitive user interface that makes automated tracking accessible to researchers with no image-processing experience or familiarity with particle-tracking approaches. Using this GUI, users can interactively determine a minimum of four parameters to identify fluorescently labeled cells and automate acquisition of cell trajectories. Additional features allow for batch processing of numerous time-lapse images, curation of unwanted tracks, and subsequent statistical analysis of tracked cells. Statistical outputs allow users to evaluate migratory phenotypes, including cell speed, distance, displacement, and persistence, as well as measures of directional movement, such as forward migration index (FMI) and angular displacement. © 2017 by John Wiley & Sons, Inc.
Keywords: automated tracking; cell migration; cell tracking; time-lapse imaging.
Copyright © 2017 John Wiley & Sons, Inc.
Figures










References
-
- Crocker JC, Grier DG. Methods of digital video microscopy for colloidal studies. J Colloid Interf Sci. 1996;179:298–310.
-
- Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. - PubMed
-
- Hand AJ, Sun T, Barber DC, Hose DR, MacNeil S. Automated tracking of migrating cells in phase-contrast video microscopy sequences using image registration. J Microsc. 2009;234:62–79. - PubMed
-
- Manzo C, Garcia-Parajo MF. A review of progress in single particle tracking: from methods to biophysical insights. Rep Prog Phys. 2015;78 - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

