A new and updated resource for codon usage tables
- PMID: 28865429
- PMCID: PMC5581930
- DOI: 10.1186/s12859-017-1793-7
A new and updated resource for codon usage tables
Abstract
Background: Due to the degeneracy of the genetic code, most amino acids can be encoded by multiple synonymous codons. Synonymous codons naturally occur with different frequencies in different organisms. The choice of codons may affect protein expression, structure, and function. Recombinant gene technologies commonly take advantage of the former effect by implementing a technique termed codon optimization, in which codons are replaced with synonymous ones in order to increase protein expression. This technique relies on the accurate knowledge of codon usage frequencies. Accurately quantifying codon usage bias for different organisms is useful not only for codon optimization, but also for evolutionary and translation studies: phylogenetic relations of organisms, and host-pathogen co-evolution relationships, may be explored through their codon usage similarities. Furthermore, codon usage has been shown to affect protein structure and function through interfering with translation kinetics, and cotranslational protein folding.
Results: Despite the obvious need for accurate codon usage tables, currently available resources are either limited in scope, encompassing only organisms from specific domains of life, or greatly outdated. Taking advantage of the exponential growth of GenBank and the creation of NCBI's RefSeq database, we have developed a new database, the High-performance Integrated Virtual Environment-Codon Usage Tables (HIVE-CUTs), to present and analyse codon usage tables for every organism with publicly available sequencing data. Compared to existing databases, this new database is more comprehensive, addresses concerns that limited the accuracy of earlier databases, and provides several new functionalities, such as the ability to view and compare codon usage between individual organisms and across taxonomical clades, through graphical representation or through commonly used indices. In addition, it is being routinely updated to keep up with the continuous flow of new data in GenBank and RefSeq.
Conclusion: Given the impact of codon usage bias on recombinant gene technologies, this database will facilitate effective development and review of recombinant drug products and will be instrumental in a wide area of biological research. The database is available at hive.biochemistry.gwu.edu/review/codon .
Keywords: Codon optimization; Codon usage bias; Recombinant protein therapeutics; Translational kinetics.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
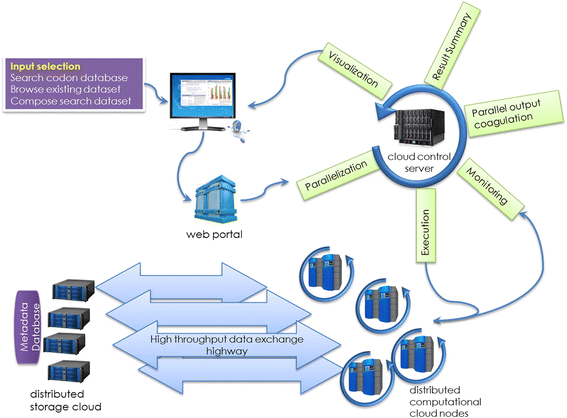
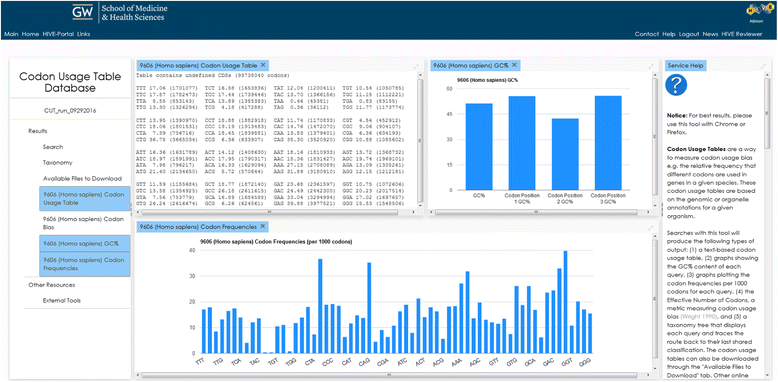
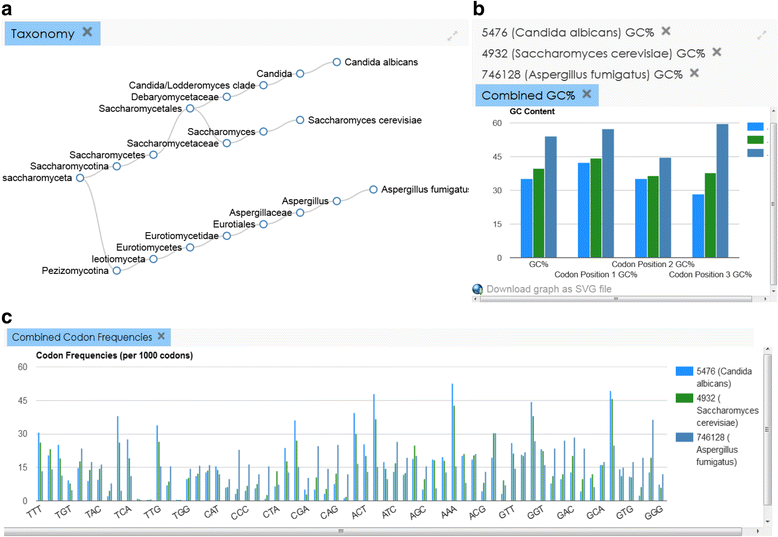
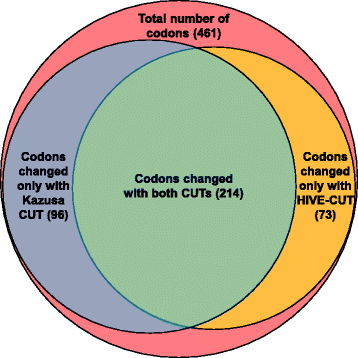
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

