Missense variants in the chromatin remodeler CHD1 are associated with neurodevelopmental disability
- PMID: 28866611
- PMCID: PMC5834353
- DOI: 10.1136/jmedgenet-2017-104759
Missense variants in the chromatin remodeler CHD1 are associated with neurodevelopmental disability
Abstract
Background: The list of Mendelian disorders of the epigenetic machinery has expanded rapidly during the last 5 years. A few missense variants in the chromatin remodeler CHD1 have been found in several large-scale sequencing efforts focused on uncovering the genetic aetiology of autism.
Objectives: To explore whether variants in CHD1 are associated with a human phenotype.
Methods: We used GeneMatcher to identify other physicians caring for patients with variants in CHD1. We also explored the epigenetic consequences of one of these variants in cultured fibroblasts.
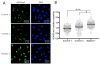
Results: Here we describe six CHD1 heterozygous missense variants in a cohort of patients with autism, speech apraxia, developmental delay and facial dysmorphic features. Importantly, three of these variants occurred de novo. We also report on a subject with a de novo deletion covering a large fraction of the CHD1 gene without any obvious neurological phenotype. Finally, we demonstrate increased levels of the closed chromatin modification H3K27me3 in fibroblasts from a subject carrying a de novo variant in CHD1.
Conclusions: Our results suggest that variants in CHD1 can lead to diverse phenotypic outcomes; however, the neurodevelopmental phenotype appears to be limited to patients with missense variants, which is compatible with a dominant negative mechanism of disease.
Keywords: chromatin; epigenetic machinery; human disease; neurological dysfunction; speech apraxia.
© Article author(s) (or their employer(s) unless otherwise stated in the text of the article) 2018. All rights reserved. No commercial use is permitted unless otherwise expressly granted.
Conflict of interest statement
Competing interests: None declared.
Figures

References
-
- Vissers LE, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17(1):9–18. - PubMed
-
- Law MJ, Lower KM, Voon HP, Hughes JR, Garrick D, Viprakasit V, Mitson M, De Gobbi M, Marra M, Morris A, Abbott A, Wilder SP, Taylor S, Santos GM, Cross J, Ayyub H, Jones S, Ragoussis J, Rhodes D, Dunham I, Higgs DR, Gibbons RJ. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143(3):367–78. - PubMed
-
- Weiss K, Terhal PA, Cohen L, Bruccoleri M, Irving M, Martinez AF, Rosenfeld JA, Machol K, Yang Y, Liu P, Walkiewicz M, Beuten J, Gomez-Ospina N, Haude K, Fong CT, Enns GM, Bernstein JA, Fan J, Gotway G, Ghorbani M, Study DDD, van Gassen K, Monroe GR, van Haaften G, Basel-Vanagaite L, Yang XJ, Campeau PM, Muenke M. De Novo Mutations in CHD4, an ATP-Dependent Chromatin Remodeler Gene, Cause an Intellectual Disability Syndrome with Distinctive Dysmorphisms. Am J Hum Genet. 2016;99(4):934–41. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
