Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis
- PMID: 28868132
- PMCID: PMC5553090
- DOI: 10.12688/f1000research.10571.2
Comprehensive comparison of Pacific Biosciences and Oxford Nanopore Technologies and their applications to transcriptome analysis
Abstract
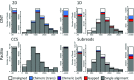
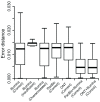
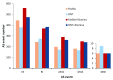
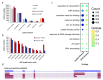
Background: Given the demonstrated utility of Third Generation Sequencing [Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT)] long reads in many studies, a comprehensive analysis and comparison of their data quality and applications is in high demand. Methods: Based on the transcriptome sequencing data from human embryonic stem cells, we analyzed multiple data features of PacBio and ONT, including error pattern, length, mappability and technical improvements over previous platforms. We also evaluated their application to transcriptome analyses, such as isoform identification and quantification and characterization of transcriptome complexity, by comparing the performance of size-selected PacBio, non-size-selected ONT and their corresponding Hybrid-Seq strategies (PacBio+Illumina and ONT+Illumina). Results: PacBio shows overall better data quality, while ONT provides a higher yield. As with data quality, PacBio performs marginally better than ONT in most aspects for both long reads only and Hybrid-Seq strategies in transcriptome analysis. In addition, Hybrid-Seq shows superior performance over long reads only in most transcriptome analyses. Conclusions: Both PacBio and ONT sequencing are suitable for full-length single-molecule transcriptome analysis. As this first use of ONT reads in a Hybrid-Seq analysis has shown, both PacBio and ONT can benefit from a combined Illumina strategy. The tools and analytical methods developed here provide a resource for future applications and evaluations of these rapidly-changing technologies.
Keywords: Oxford Nanopore Technologies; PacBio; Third Generation Sequencing; Transcriptome.
Conflict of interest statement
Competing interests: No competing interests were disclosed.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

