Improved deconvolution of very weak confocal signals
- PMID: 28868135
- PMCID: PMC5553083
- DOI: 10.12688/f1000research.11773.2
Improved deconvolution of very weak confocal signals
Abstract
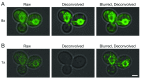
Deconvolution is typically used to sharpen fluorescence images, but when the signal-to-noise ratio is low, the primary benefit is reduced noise and a smoother appearance of the fluorescent structures. 3D time-lapse (4D) confocal image sets can be improved by deconvolution. However, when the confocal signals are very weak, the popular Huygens deconvolution software erases fluorescent structures that are clearly visible in the raw data. We find that this problem can be avoided by prefiltering the optical sections with a Gaussian blur. Analysis of real and simulated data indicates that the Gaussian blur prefilter preserves meaningful signals while enabling removal of background noise. This approach is very simple, and it allows Huygens to be used with 4D imaging conditions that minimize photodamage.
Keywords: 4D microscopy; Gaussian blur; Huygens; confocal microscopy; deconvolution; fluorescence microscopy; signal-to-noise.
Conflict of interest statement
Competing interests: No competing interests were disclosed.
Figures



References
-
- Burger W, Burge MJ: Digital Image Processing: An Algorithmic Introduction using Java. Springer, New York NY.2008. 10.1007/978-1-84628-968-2 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

