Understanding molecular interactions between scavenger receptor A and its natural product inhibitors through molecular modeling studies
- PMID: 28869863
- PMCID: PMC5696021
- DOI: 10.1016/j.jmgm.2017.08.013
Understanding molecular interactions between scavenger receptor A and its natural product inhibitors through molecular modeling studies
Abstract
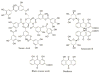
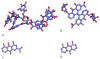
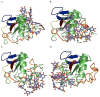
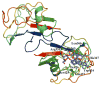
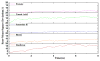
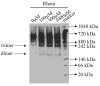
Scavenger receptor A (SRA), as an immune regulator, has been shown to play important roles in lipid metabolism, cardiovascular diseases, and pathogen recognition. Several natural product inhibitors of SRA have been studied for their potential application in modulating SRA functions. To understand the binding mode of these inhibitors on SRA, we conducted systematic molecular modeling studies in order to identify putative binding domain(s) that may be responsible for their recognition to the receptor as well as their inhibitory activity. Treatment of SRA with one of the natural product inhibitors, rhein, led to significant dissociation of SRA oligomers to its trimer and dimer forms, which further supported our hypothesis on their putative mechanism of action. Such information is believed to shed light on design of more potent inhibitors for the receptor in order to develop potential therapeutics through immune system modulation.
Keywords: Binding modes; Docking; Inhibitor; Molecular dynamics simulation; Natural products; SRA.
Published by Elsevier Inc.
Figures
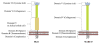






References
-
- Platt N, Haworth R, Darley L, Gordon S. The many roles of the class A macrophage scavenger receptor. International Review of Cytology. 2002;212:1–40. - PubMed
-
- Freeman M, Ashkenes J, Rees DJ, Kingsley DM, Copeland NG, Jenkins NA, Krieger M. An ancient, highly conserved family of cysteine-rich protein domains revealed by cloning type I and type II murine macrophage scavenger receptors. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:8810–8814. - PMC - PubMed
-
- Suzuki H, Kurihara Y, Takeya M, Kamada N, Kataoka M, Jishage K, Ueda O, Sakaguchi H, Higashi T, Suzuki T, Takashima Y, Kawabe Y, Cynshi O, Wada Y, Honda M, Kurihara H, Aburatani H, Doi T, Matsumoto A, Azuma S, et al. A role of macrophage scavenger receptors in atherosclerosis and susceptibility to infection. Nature. 1997;386:292–296. - PubMed
-
- Ricci R, Sumara G, Sumara I, Rozenberg I, Kurrer M, Akhmedov A, Hersberger M, Eriksson U, Eberli FR, Becher B, Boren J, Chen M, Cybulsky MI, Moore KJ, Freeman MW, Wagner EF, Matter CM, Luscher TF. Requirement of JNK2 for scavenger receptor A-mediated foam cell formation in atherogenesis. Science. 2004;306:1558–1561. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

