Palmitoylation-dependent activation of MC1R prevents melanomagenesis
- PMID: 28869973
- PMCID: PMC5902815
- DOI: 10.1038/nature23887
Palmitoylation-dependent activation of MC1R prevents melanomagenesis
Abstract
The melanocortin-1 receptor (MC1R), a G-protein-coupled receptor, has a crucial role in human and mouse pigmentation. Activation of MC1R in melanocytes by α-melanocyte-stimulating hormone (α-MSH) stimulates cAMP signalling and melanin production and enhances DNA repair after ultraviolet irradiation. Individuals carrying MC1R variants, especially those associated with red hair colour, fair skin and poor tanning ability (denoted as RHC variants), are associated with higher risk of melanoma. However, how MC1R activity is modulated by ultraviolet irradiation, why individuals with red hair are more prone to developing melanoma, and whether the activity of RHC variants might be restored for therapeutic benefit are unknown. Here we demonstrate a potential MC1R-targeted intervention strategy in mice to rescue loss-of-function MC1R in MC1R RHC variants for therapeutic benefit by activating MC1R protein palmitoylation. MC1R palmitoylation, primarily mediated by the protein-acyl transferase ZDHHC13, is essential for activating MC1R signalling, which triggers increased pigmentation, ultraviolet-B-induced G1-like cell cycle arrest and control of senescence and melanomagenesis in vitro and in vivo. Using C57BL/6J-Mc1re/eJ mice, in which endogenous MC1R is prematurely terminated, expressing Mc1r RHC variants, we show that pharmacological activation of palmitoylation rescues the defects of Mc1r RHC variants and prevents melanomagenesis. The results highlight a central role for MC1R palmitoylation in pigmentation and protection against melanoma.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
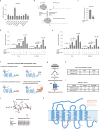


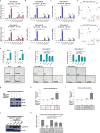

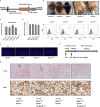


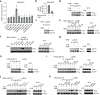


Comment in
-
Cell signalling: Red alert about lipid's role in skin cancer.Nature. 2017 Sep 21;549(7672):337-339. doi: 10.1038/nature23550. Epub 2017 Sep 6. Nature. 2017. PMID: 28869972 No abstract available.
-
Sticky fingers at work: Palmitoylation-dependent MC1R activation.Pigment Cell Melanoma Res. 2018 Mar;31(2):238-240. doi: 10.1111/pcmr.12659. Epub 2017 Nov 2. Pigment Cell Melanoma Res. 2018. PMID: 29044987 No abstract available.
References
-
- Jackson IJ, Budd PS, Keighren M, McKie L. Humanized MC1R transgenic mice reveal human specific receptor function. Hum Mol Genet. 2007;16:2341–2348. - PubMed
-
- D’Orazio JA, et al. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–344. - PubMed
-
- Healy E, et al. Functional variation of MC1R alleles from red-haired individuals. Hum Mol Genet. 2001;10:2397–2402. - PubMed
-
- Raimondi S, et al. MC1R variants, melanoma and red hair color phenotype: a meta-analysis. Int J Cancer. 2008;122:2753–2760. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

