Dual RNA-seq reveals no plastic transcriptional response of the coccidian parasite Eimeria falciformis to host immune defenses
- PMID: 28870168
- PMCID: PMC5584376
- DOI: 10.1186/s12864-017-4095-6
Dual RNA-seq reveals no plastic transcriptional response of the coccidian parasite Eimeria falciformis to host immune defenses
Abstract
Background: Parasites can either respond to differences in immune defenses that exist between individual hosts plastically or, alternatively, follow a genetically canalized ("hard wired") program of infection. Assuming that large-scale functional plasticity would be discernible in the parasite transcriptome we have performed a dual RNA-seq study of the lifecycle of Eimeria falciformis using infected mice with different immune status as models for coccidian infections.
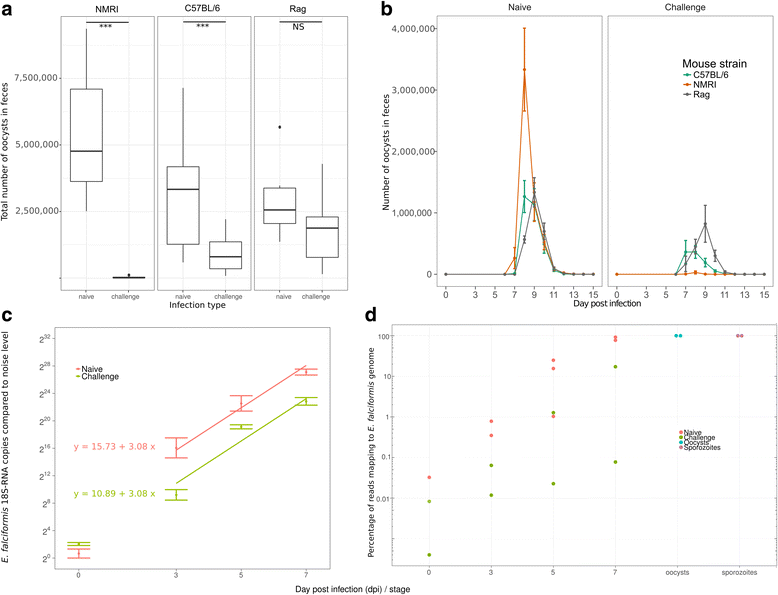
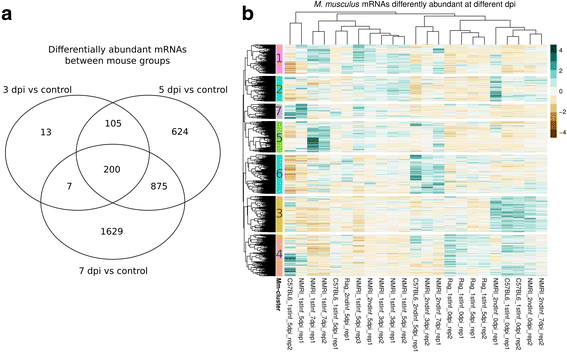
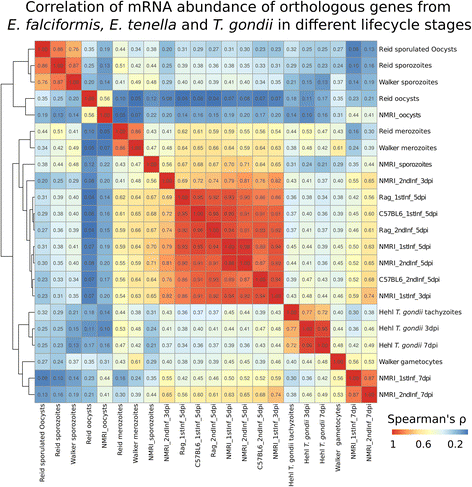
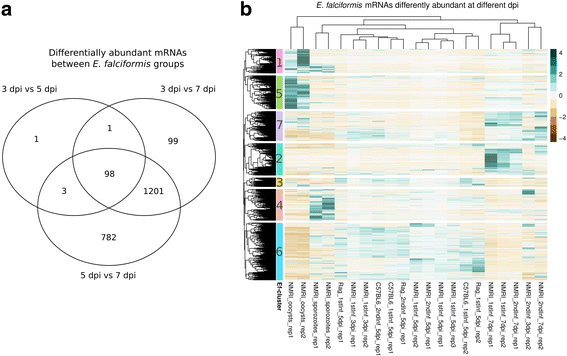
Results: We compared parasite and host transcriptomes (dual transcriptome) between naïve and challenge infected mice, as well as between immune competent and immune deficient ones. Mice with different immune competence show transcriptional differences as well as differences in parasite reproduction (oocyst shedding). Broad gene categories represented by differently abundant host genes indicate enrichments for immune reaction and tissue repair functions. More specifically, TGF-beta, EGF, TNF and IL-1 and IL-6 are examples of functional annotations represented differently depending on host immune status. Much in contrast, parasite transcriptomes were neither different between Coccidia isolated from immune competent and immune deficient mice, nor between those harvested from naïve and challenge infected mice. Instead, parasite transcriptomes have distinct profiles early and late in infection, characterized largely by biosynthesis or motility associated functional gene groups, respectively. Extracellular sporozoite and oocyst stages showed distinct transcriptional profiles and sporozoite transcriptomes were found enriched for species specific genes and likely pathogenicity factors.
Conclusion: We propose that the niche and host-specific parasite E. falciformis uses a genetically canalized program of infection. This program is likely fixed in an evolutionary process rather than employing phenotypic plasticity to interact with its host. This in turn might limit the potential of the parasite to adapt to new host species or niches, forcing it to coevolve with its host.
Keywords: Apicomplexa; Coccidia; Dual RNA-seq; Dual transcriptomics; Parasite lifecycle; Phenotypic plasticity; Transcriptional plasticity.
Conflict of interest statement
Ethics approval and consent to participate
Animal procedures were performed according to the German Animal Protection Laws as directed and approved by the overseeing authority Landesamt fuer Gesundheit und Soziales (Berlin, Germany) under numbers H0098/04 and G0039/11.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Stearns SC. The evolutionary significance of phenotypic plasticity. Bioscience. 1989;39:436–445. doi: 10.2307/1311135. - DOI
-
- Dodson S. Predator-induced reaction norms. Bioscience. 1989;39:447–452. doi: 10.2307/1311136. - DOI
-
- Stear MJ, Bairden K, Duncan JL, Holmes PH, McKellar QA, Park M, et al. How hosts control worms. Nature 1997;389:27–27. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

