The lysin motif-containing proteins, Lyp1, Lyk7 and LysMe3, play important roles in chitin perception and defense against Verticillium dahliae in cotton
- PMID: 28870172
- PMCID: PMC5583995
- DOI: 10.1186/s12870-017-1096-1
The lysin motif-containing proteins, Lyp1, Lyk7 and LysMe3, play important roles in chitin perception and defense against Verticillium dahliae in cotton
Abstract
Background: Lysin motif (LysM)-containing proteins are important pattern recognition receptors (PRRs) in plants, which function in the perception of microbe-associated molecular patterns (MAMPs) and in the defense against pathogenic attack. To date, the LysM genes have not been systematically analyzed in cotton or effectively utilized for disease resistance.
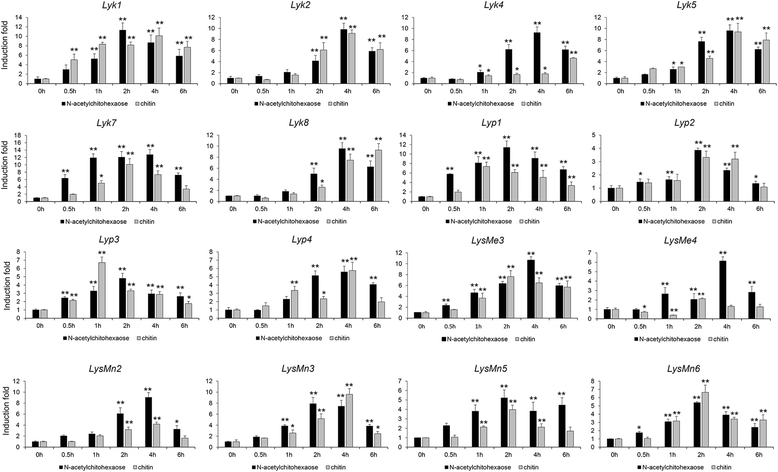
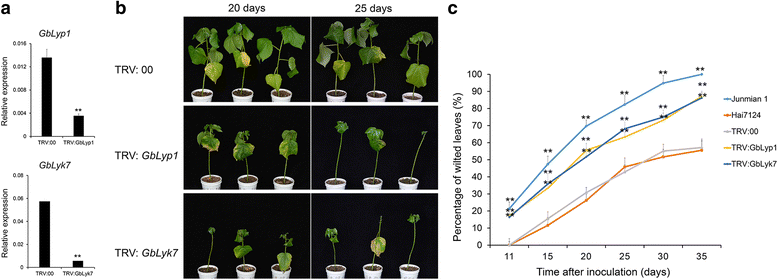
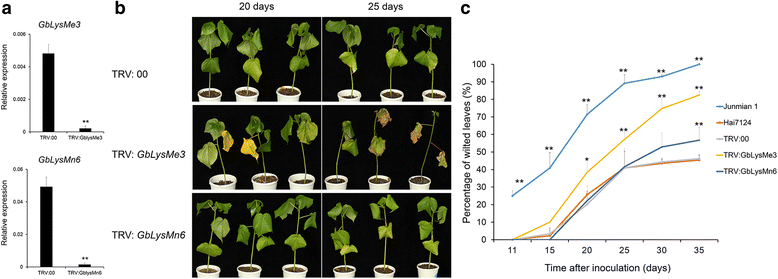
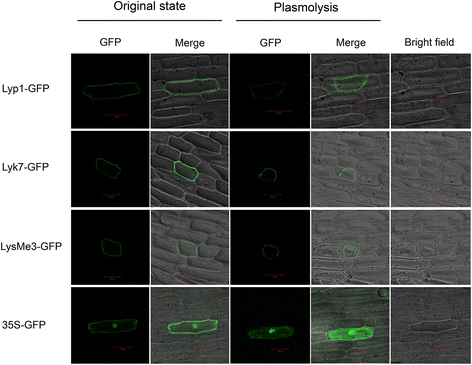
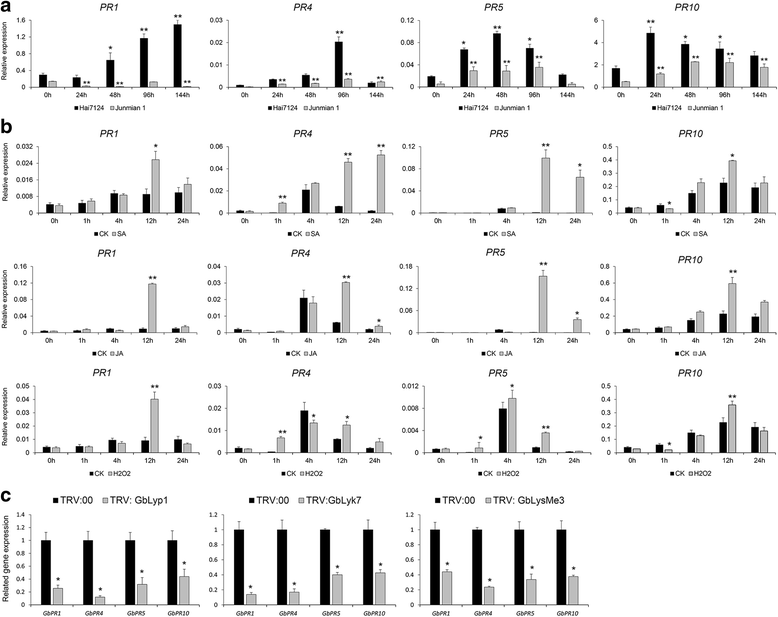
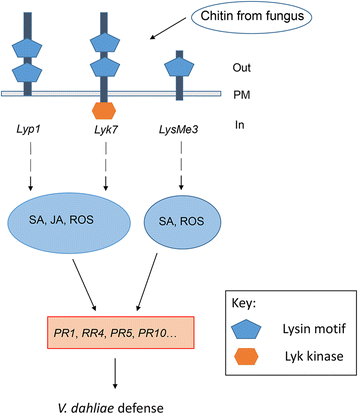
Results: Here, we identified 29, 30, 60, and 56 LysM genes in the four sequenced cotton species, diploid Gossypium raimondii, diploid G. arboreum, tetraploid G. hirsutum acc. TM-1, and G. barbadense acc. 3-79, respectively. These LysM genes were classified into four groups with different structural characteristics and a variety of expression patterns in different organs and tissues when induced by chitin or Verticillium dahliae. We further characterized three genes, Lyp1, Lyk7 and LysMe3, which showed significant increase in expression in response to chitin signals, V. dahliae challenge and several stress-related signaling compounds. Lyp1, Lyk7 and LysMe3 proteins were localized to the plasma membrane, and silencing of their expression in cotton drastically impaired salicylic acid, jasmonic acid, and reactive oxygen species generation, impaired defense gene activation, and compromised resistance to V. dahliae.
Conclusion: Our results indicate that Lyp1, Lyk7, and LysMe3 are important PRRs that function in the recognition of chitin signals to activate the downstream defense processes and induce cotton defense mechanisms against V. dahliae.
Keywords: Chitin signals; Cotton; Lysin motif; Pattern recognition receptors; Plasma membrane localization; Verticillium dahliae.
Conflict of interest statement
Ethics approval and consent to participate
Three cotton materials, including
Consent for publication
Not applicable.
Competing interests
The authors declared that they had no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




Similar articles
-
The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection.J Exp Bot. 2016 Mar;67(6):1935-50. doi: 10.1093/jxb/erw016. Epub 2016 Feb 11. J Exp Bot. 2016. PMID: 26873979 Free PMC article.
-
Proteomic and virus-induced gene silencing (VIGS) Analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae.Mol Cell Proteomics. 2013 Dec;12(12):3690-703. doi: 10.1074/mcp.M113.031013. Epub 2013 Sep 9. Mol Cell Proteomics. 2013. PMID: 24019146 Free PMC article.
-
Discovery and identification of candidate genes from the chitinase gene family for Verticillium dahliae resistance in cotton.Sci Rep. 2016 Jun 29;6:29022. doi: 10.1038/srep29022. Sci Rep. 2016. PMID: 27354165 Free PMC article.
-
Insights to Gossypium defense response against Verticillium dahliae: the Cotton Cancer.Funct Integr Genomics. 2023 May 1;23(2):142. doi: 10.1007/s10142-023-01065-5. Funct Integr Genomics. 2023. PMID: 37121989 Review.
-
Physiological and molecular mechanism of defense in cotton against Verticillium dahliae.Plant Physiol Biochem. 2018 Apr;125:193-204. doi: 10.1016/j.plaphy.2018.02.011. Epub 2018 Feb 13. Plant Physiol Biochem. 2018. PMID: 29462745 Review.
Cited by
-
An Overview of the Molecular Genetics of Plant Resistance to the Verticillium Wilt Pathogen Verticillium dahliae.Int J Mol Sci. 2020 Feb 7;21(3):1120. doi: 10.3390/ijms21031120. Int J Mol Sci. 2020. PMID: 32046212 Free PMC article. Review.
-
Host-induced gene silencing of a regulator of G protein signalling gene (VdRGS1) confers resistance to Verticillium wilt in cotton.Plant Biotechnol J. 2018 Feb 12;16(9):1629-43. doi: 10.1111/pbi.12900. Online ahead of print. Plant Biotechnol J. 2018. PMID: 29431919 Free PMC article.
-
Transcriptome analysis of Gossypium hirsutum cultivar Zhongzhimian No.2 uncovers the gene regulatory networks involved in defense against Verticillium dahliae.BMC Plant Biol. 2024 May 27;24(1):457. doi: 10.1186/s12870-024-05165-7. BMC Plant Biol. 2024. PMID: 38797823 Free PMC article.
-
Genetic Basis of Fiber Improvement and Decreased Stress Tolerance in Cultivated Versus Semi-Domesticated Upland Cotton.Front Plant Sci. 2019 Nov 29;10:1572. doi: 10.3389/fpls.2019.01572. eCollection 2019. Front Plant Sci. 2019. PMID: 31850042 Free PMC article.
-
RNA-Sequencing, Physiological and RNAi Analyses Provide Insights into the Response Mechanism of the ABC-Mediated Resistance to Verticillium dahliae Infection in Cotton.Genes (Basel). 2019 Feb 1;10(2):110. doi: 10.3390/genes10020110. Genes (Basel). 2019. PMID: 30717226 Free PMC article.
References
-
- Baureithel K, Felix G, Boller T. Specific, high affinity binding of chitin fragments to tomato cells and membranes. Competitive inhibition of binding by derivatives of chitooligosaccharides and a nod factor of Rhizobium. J Biol Chem. 1994;269(27):17931–17938. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

