Up-modulation of PLC-β2 reduces the number and malignancy of triple-negative breast tumor cells with a CD133+/EpCAM+ phenotype: a promising target for preventing progression of TNBC
- PMID: 28870198
- PMCID: PMC5584040
- DOI: 10.1186/s12885-017-3592-y
Up-modulation of PLC-β2 reduces the number and malignancy of triple-negative breast tumor cells with a CD133+/EpCAM+ phenotype: a promising target for preventing progression of TNBC
Abstract
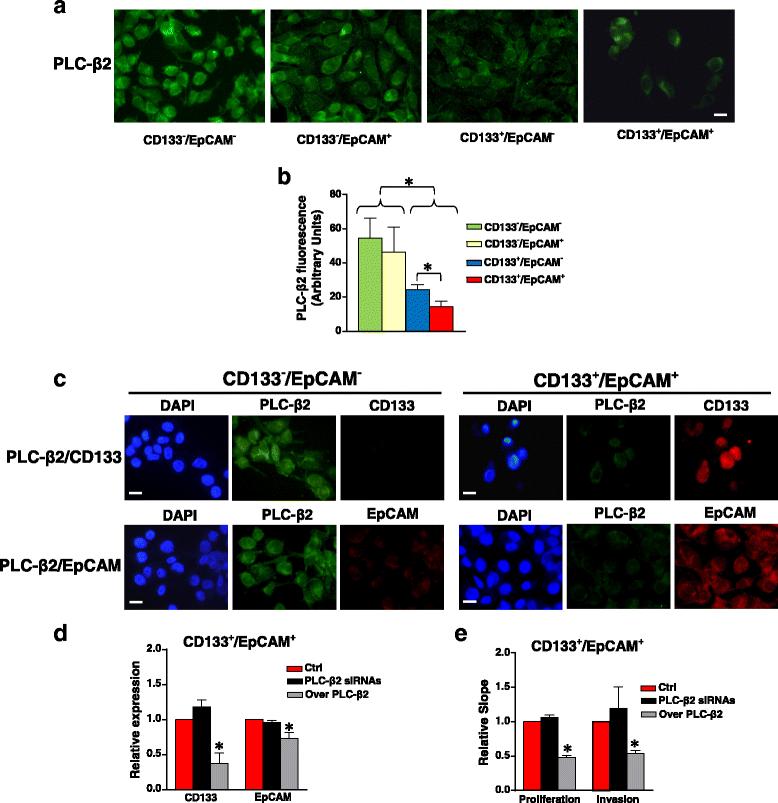
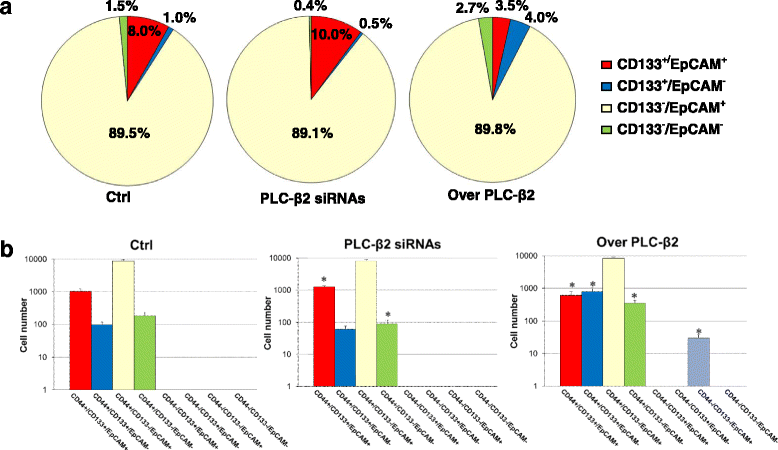
Background: The malignant potential of triple negative breast cancer (TNBC) is also dependent on a sub-population of cells with a stem-like phenotype. Among the cancer stem cell markers, CD133 and EpCAM strongly correlate with breast tumor aggressiveness, suggesting that simultaneous targeting of the two surface antigens may be beneficial in treatment of TNBC. Since in TNBC-derived cells we demonstrated that PLC-β2 induces the conversion of CD133high to CD133low cells, here we explored its possible role in down-modulating the expression of both CD133 and EpCAM and, ultimately, in reducing the number of TNBC cells with a stem-like phenotype.
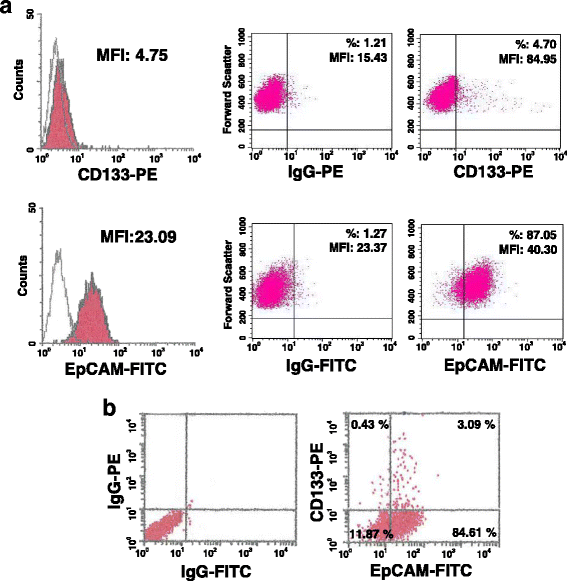
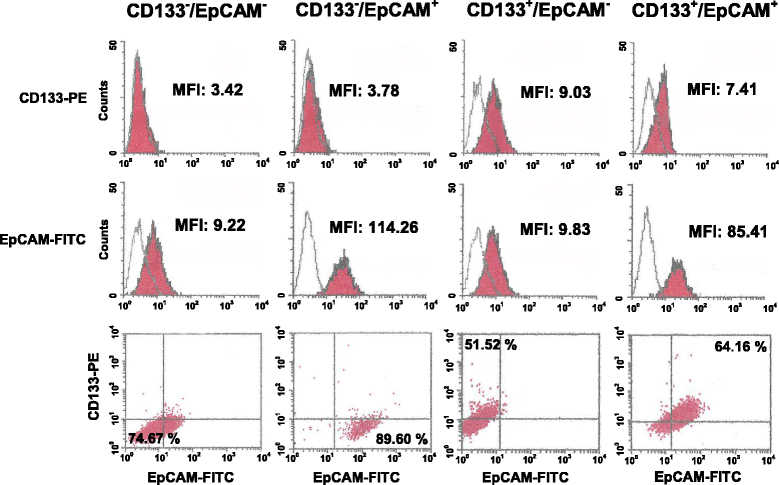
Methods: A magnetic step-by-step cell isolation with antibodies directed against CD133 and/or EpCAM was performed on the TNBC-derived MDA-MB-231 cell line. In the same cell model, PLC-β2 was over-expressed or down-modulated and cell proliferation and invasion capability were evaluated by Real-time cell assays. The surface expression of CD133, EpCAM and CD44 in the different experimental conditions were measured by multi-color flow cytometry immunophenotyping.
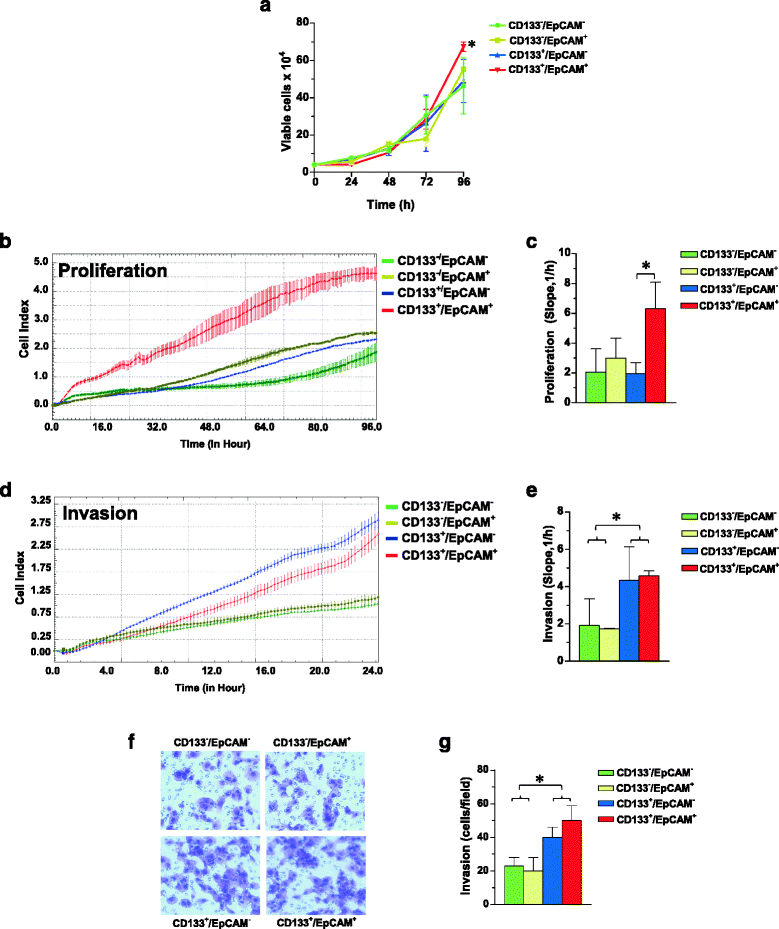
Results: A CD133+/EpCAM+ sub-population with high proliferation rate and invasion capability is present in the MDA-MB-231 cell line. Over-expression of PLC-β2 in CD133+/EpCAM+ cells reduced the surface expression of both CD133 and EpCAM, as well as proliferation and invasion capability of this cellular subset. On the other hand, the up-modulation of PLC-β2 in the whole MDA-MB-231 cell population reduced the number of cells with a CD44+/CD133+/EpCAM+ stem-like phenotype.
Conclusions: Since selective targeting of the cells with the highest aggressive potential may have a great clinical importance for TNBC, the up-modulation of PLC-β2, reducing the number of cells with a stem-like phenotype, may be a promising goal for novel therapies aimed to prevent the progression of aggressive breast tumors.
Keywords: Breast cancer stem cell (BCSC); CD133; EpCAM; Invasiveness; PLC-β2; Proliferation; Triple-negative breast cancer (TNBC).
Conflict of interest statement
Authors’ information
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
In triple negative breast tumor cells, PLC-β2 promotes the conversion of CD133high to CD133low phenotype and reduces the CD133-related invasiveness.Mol Cancer. 2013 Dec 13;12:165. doi: 10.1186/1476-4598-12-165. Mol Cancer. 2013. PMID: 24330829 Free PMC article.
-
PLC-β2 is modulated by low oxygen availability in breast tumor cells and plays a phenotype dependent role in their hypoxia-related malignant potential.Mol Carcinog. 2016 Dec;55(12):2210-2221. doi: 10.1002/mc.22462. Epub 2016 Jan 19. Mol Carcinog. 2016. PMID: 26785288
-
Granzyme B-based cytolytic fusion protein targeting EpCAM specifically kills triple negative breast cancer cells in vitro and inhibits tumor growth in a subcutaneous mouse tumor model.Cancer Lett. 2016 Mar 28;372(2):201-9. doi: 10.1016/j.canlet.2016.01.027. Epub 2016 Jan 21. Cancer Lett. 2016. PMID: 26806809
-
Cancer stem cells in triple-negative breast cancer: a potential target and prognostic marker.Biomark Med. 2018 Jul;12(7):813-820. doi: 10.2217/bmm-2017-0398. Epub 2018 Jun 15. Biomark Med. 2018. PMID: 29902924 Review.
-
Ganglioside GD2: a novel therapeutic target in triple-negative breast cancer.Ann N Y Acad Sci. 2022 Feb;1508(1):35-53. doi: 10.1111/nyas.14700. Epub 2021 Oct 1. Ann N Y Acad Sci. 2022. PMID: 34596246 Review.
Cited by
-
Microfluidic Isolation of Circulating Tumor Cells and Cancer Stem-Like Cells from Patients with Pancreatic Ductal Adenocarcinoma.Theranostics. 2019 Feb 20;9(5):1417-1425. doi: 10.7150/thno.28745. eCollection 2019. Theranostics. 2019. PMID: 30867841 Free PMC article.
-
Ectopic expression of PLC-β2 in non-invasive breast tumor cells plays a protective role against malignant progression and is correlated with the deregulation of miR-146a.Mol Carcinog. 2019 May;58(5):708-721. doi: 10.1002/mc.22964. Epub 2019 Jan 16. Mol Carcinog. 2019. PMID: 30582225 Free PMC article.
-
Adhesion to the Brain Endothelium Selects Breast Cancer Cells with Brain Metastasis Potential.Int J Mol Sci. 2023 Apr 11;24(8):7087. doi: 10.3390/ijms24087087. Int J Mol Sci. 2023. PMID: 37108248 Free PMC article.
-
Molecular regulation of PLCβ signaling.Methods Enzymol. 2023;682:17-52. doi: 10.1016/bs.mie.2023.01.001. Epub 2023 Feb 22. Methods Enzymol. 2023. PMID: 36948701 Free PMC article. Review.
-
CD133 mRNA-transfected dendritic cells induce coordinated cytotoxic and helper T cell responses against breast cancer stem cells.Mol Ther Oncolytics. 2021 May 19;22:64-71. doi: 10.1016/j.omto.2021.05.006. eCollection 2021 Sep 24. Mol Ther Oncolytics. 2021. PMID: 34485687 Free PMC article.
References
-
- Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, Savage MI, Osborne CK, Hilsenbeck SG, Chang JC, Mills GB, Lau CC, Brown PH. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res. 2015;21:1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. - DOI - PMC - PubMed
-
- Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, Brown M, Viens P, Xerri L, Bertucci F, Stassi G, Dontu G, Birnbaum D, Wicha MS. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

