Engineering a riboswitch-based genetic platform for the self-directed evolution of acid-tolerant phenotypes
- PMID: 28871084
- PMCID: PMC5583362
- DOI: 10.1038/s41467-017-00511-w
Engineering a riboswitch-based genetic platform for the self-directed evolution of acid-tolerant phenotypes
Abstract
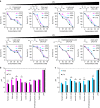
Environmental pH is a fundamental signal continuously directing the metabolism and behavior of living cells. Programming the precise cellular response toward environmental pH is, therefore, crucial for engineering cells for increasingly sophisticated functions. Herein, we engineer a set of riboswitch-based pH-sensing genetic devices to enable the control of gene expression according to differential environmental pH. We next develop a digital pH-sensing system to utilize the analogue-sensing behavior of these devices for high-resolution recording of host cell exposure to discrete external pH levels. The application of this digital pH-sensing system is demonstrated in a genetic program that autonomously regulated the evolutionary engineering of host cells for improved tolerance to a broad spectrum of organic acids, a valuable phenotype for metabolic engineering and bioremediation applications.Cells are exposed to shifts in environmental pH, which direct their metabolism and behavior. Here the authors design pH-sensing riboswitches to create a gene expression program, digitalize the system to respond to a narrow pH range and apply it to evolve host cells with improved tolerance to a variety of organic acids.
Conflict of interest statement
A patent application related to some of the work presented here has been filed on behalf of the National University of Singapore.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

