TRIM23 mediates virus-induced autophagy via activation of TBK1
- PMID: 28871090
- PMCID: PMC5658249
- DOI: 10.1038/s41564-017-0017-2
TRIM23 mediates virus-induced autophagy via activation of TBK1
Abstract
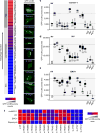
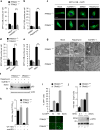
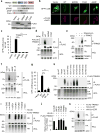
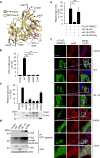
Autophagy and interferon (IFN)-mediated innate immunity are critical antiviral defence mechanisms, and recent evidence indicated that tripartite motif (TRIM) proteins are important regulators of both processes. Although the role of TRIM proteins in modulating antiviral cytokine responses has been well established, much less is known about their involvement in autophagy in response to different viral pathogens. Through a targeted RNAi screen examining the relevance of selected TRIM proteins in autophagy induced by herpes simplex virus 1 (HSV-1), encephalomyocarditis virus (EMCV) and influenza A virus (IAV), we identified several TRIM proteins that regulate autophagy in a virus-species-specific manner, as well as a few TRIM proteins that were essential for autophagy triggered by all three viruses and rapamycin, among them TRIM23. TRIM23 was critical for autophagy-mediated restriction of multiple viruses, and this activity was dependent on both its RING E3 ligase and ADP-ribosylation factor (ARF) GTPase activity. Mechanistic studies revealed that unconventional K27-linked auto-ubiquitination of the ARF domain is essential for the GTP hydrolysis activity of TRIM23 and activation of TANK-binding kinase 1 (TBK1) by facilitating its dimerization and ability to phosphorylate the selective autophagy receptor p62. Our work identifies the TRIM23-TBK1-p62 axis as a key component of selective autophagy and further reveals a role for K27-linked ubiquitination in GTPase-dependent TBK1 activation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

