Microbial Diversity in Sulfate-Reducing Marine Sediment Enrichment Cultures Associated with Anaerobic Biotransformation of Coastal Stockpiled Phosphogypsum (Sfax, Tunisia)
- PMID: 28871244
- PMCID: PMC5566975
- DOI: 10.3389/fmicb.2017.01583
Microbial Diversity in Sulfate-Reducing Marine Sediment Enrichment Cultures Associated with Anaerobic Biotransformation of Coastal Stockpiled Phosphogypsum (Sfax, Tunisia)
Abstract
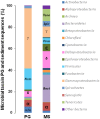
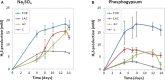
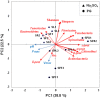
Anaerobic biotechnology using sulfate-reducing bacteria (SRB) is a promising alternative for reducing long-term stockpiling of phosphogypsum (PG), an acidic (pH ~3) by-product of the phosphate fertilizer industries containing high amounts of sulfate. The main objective of this study was to evaluate, for the first time, the diversity and ability of anaerobic marine microorganisms to convert sulfate from PG into sulfide, in order to look for marine SRB of biotechnological interest. A series of sulfate-reducing enrichment cultures were performed using different electron donors (i.e., acetate, formate, or lactate) and sulfate sources (i.e., sodium sulfate or PG) as electron acceptors. Significant sulfide production was observed from enrichment cultures inoculated with marine sediments, collected near the effluent discharge point of a Tunisian fertilizer industry (Sfax, Tunisia). Sulfate sources impacted sulfide production rates from marine sediments as well as the diversity of SRB species belonging to Deltaproteobacteria. When PG was used as sulfate source, Desulfovibrio species dominated microbial communities of marine sediments, while Desulfobacter species were mainly detected using sodium sulfate. Sulfide production was also affected depending on the electron donor used, with the highest production obtained using formate. In contrast, low sulfide production (acetate-containing cultures) was associated with an increase in the population of Firmicutes. These results suggested that marine Desulfovibrio species, to be further isolated, are potential candidates for bioremediation of PG by immobilizing metals and metalloids thanks to sulfide production by these SRB.
Keywords: Desulfovibrio; anaerobes; anaerobic biotechnology; bioremediation; marine sediment; next-generation sequencing; phosphogypsum; sulfate-reducing bacteria.
Figures



References
-
- Ajam L., Ouezdou M. B., Felfoul H. S., El Mensi R. (2009). Characterization of the Tunisian phosphogypsum and its valorization in clay bricks. Constr. Build. Mater. 23, 3240–3247. 10.1016/j.conbuildmat.2009.05.009 - DOI
-
- Azabou S., Mechichi T., Sayadi S. (2005). Sulfate reduction from phosphogypsum using a mixed culture of sulfate-reducing bacteria. Int. Biodeterior. Biodegradation. 56, 236–242 10.1016/j.ibiod.2005.09.003 - DOI
-
- Azabou S., Mechichi T., Sayadi S. (2007a). Zinc precipitation by heavy-metal tolerant sulfate-reducing bacteria enriched on phosphogypsum as a sulfate source. Miner. Eng. 20, 173–178. 10.1016/j.mineng.2006.08.008 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

