Succession and Fermentation Products of Grass Carp (Ctenopharyngodon idellus) Hindgut Microbiota in Response to an Extreme Dietary Shift
- PMID: 28871246
- PMCID: PMC5566599
- DOI: 10.3389/fmicb.2017.01585
Succession and Fermentation Products of Grass Carp (Ctenopharyngodon idellus) Hindgut Microbiota in Response to an Extreme Dietary Shift
Abstract
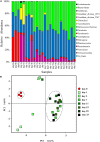
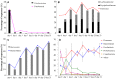
Dietary intake affects the structure and function of microbes in host intestine. However, the succession of gut microbiota in response to changes in macronutrient levels during a long period of time remains insufficiently studied. Here, we determined the succession and metabolic products of intestinal microbiota in grass carp (Ctenopharyngodon idellus) undergoing an abrupt and extreme diet change, from fish meal to Sudan grass (Sorghum sudanense). Grass carp hindgut microbiota responded rapidly to the diet shift, reaching a new equilibrium approximately within 11 days. In comparison to animal-diet samples, Bacteroides, Lachnospiraceae and Erysipelotrichaceae increased significantly while Cetobacterium decreased significantly in plant-diet samples. Cetobacterium was negatively correlated with Bacteroides, Lachnospiraceae and Erysipelotrichaceae, while Bacteroides was positively correlated with Lachnospiraceae. Predicted glycoside hydrolase and polysaccharide lyase genes in Bacteroides and Lachnospiraceae from the Carbohydrate-Active enZymes (CAZy) database might be involved in degradation of the plant cell wall polysaccharides. However, none of these enzymes was detected in the grass carp genome searched against dbCAN database. Additionally, a significant decrease of short chain fatty acids levels in plant-based samples was observed. Generally, our results suggest a rapid adaption of grass carp intestinal microbiota to dietary shift, and that microbiota are likely to play an indispensable role in nutrient turnover and fermentation.
Keywords: SCFAs; freshwater fish; gut microbiota; high-fiber diet; high-protein diet.
Figures


References
-
- Bergman E. N. (1990). Energy contributions of volatile fatty-acids from the gastrointestinal-tract in various species. Physiol. Rev. 70 567–590. - PubMed
-
- Biddle A., Stewart L., Blanchard J., Leschine S. (2013). Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity 5 627–640. 10.3390/d5030627 - DOI
-
- Brulc J. M., Antonopoulos D. A., Miller M. E. B., Wilson M. K., Yannarell A. C., Dinsdale E. A., et al. (2009). Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc. Natl. Acad. Sci. U.S.A. 106 1948–1953. 10.1073/pnas.0806191105 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

