A miRNA catalogue and ncRNA annotation of the short-living fish Nothobranchius furzeri
- PMID: 28874118
- PMCID: PMC5584509
- DOI: 10.1186/s12864-017-3951-8
A miRNA catalogue and ncRNA annotation of the short-living fish Nothobranchius furzeri
Erratum in
-
Correction to: A miRNA catalogue and ncRNA annotation of the short-living fish Nothobranchius furzeri.BMC Genomics. 2019 Nov 27;20(1):898. doi: 10.1186/s12864-019-6312-y. BMC Genomics. 2019. PMID: 31775605 Free PMC article.
Abstract
Background: The short-lived fish Nothobranchius furzeri is the shortest-lived vertebrate that can be cultured in captivity and was recently established as a model organism for aging research. Small non-coding RNAs, especially miRNAs, are implicated in age dependent control of gene expression.
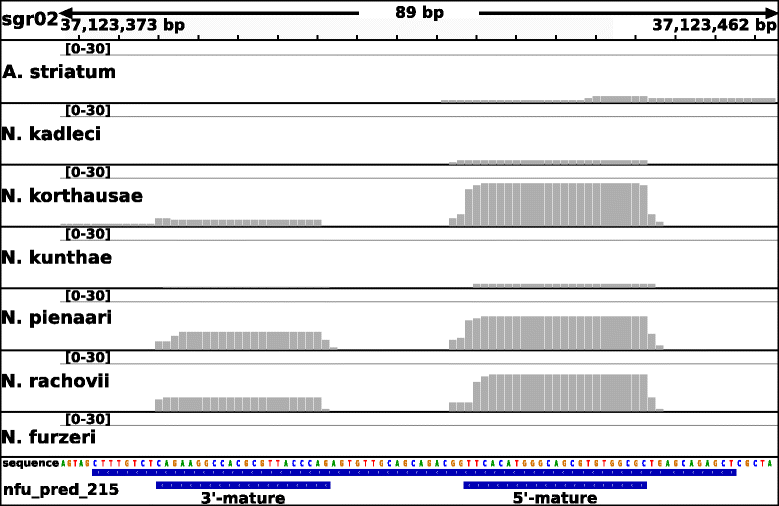
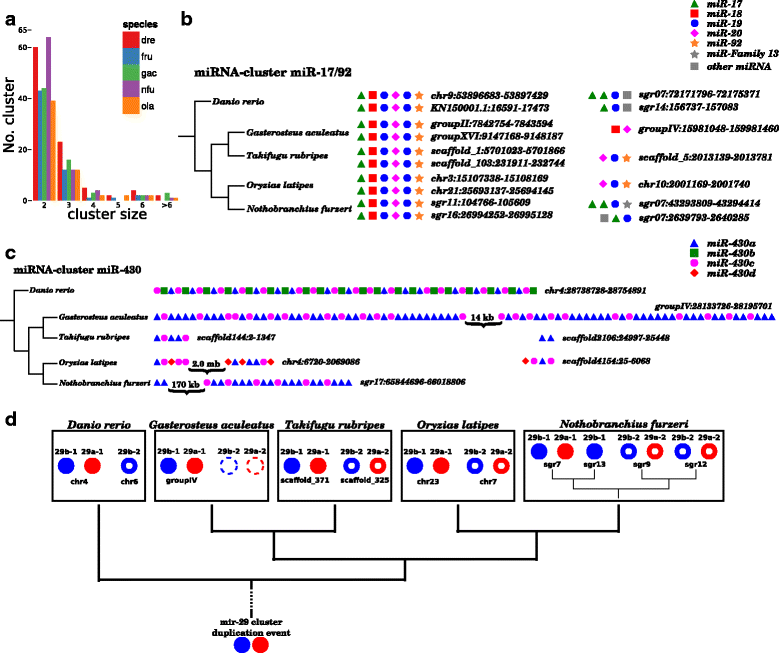
Results: Here, we present a comprehensive catalogue of miRNAs and several other non-coding RNA classes (ncRNAs) for Nothobranchius furzeri. Analyzing multiple small RNA-Seq libraries, we show most of these identified miRNAs are expressed in at least one of seven Nothobranchius species. Additionally, duplication and clustering of N. furzeri miRNAs was analyzed and compared to the four fish species Danio rerio, Oryzias latipes, Gasterosteus aculeatus and Takifugu rubripes. A peculiar characteristic of N. furzeri, as compared to other teleosts, was a duplication of the miR-29 cluster.
Conclusion: The completeness of the catalogue we provide is comparable to that of the zebrafish. This catalogue represents a basis to investigate the role of miRNAs in aging and development in this species.
Keywords: Fish miRNA evolution; Nothobranchius furzeri; miRNome; ncRNA.
Conflict of interest statement
Ethics approval and consent to participate
The protocols of animal maintenance and experiments were approved by the local authority in the State of Thuringia (Veterinaer-und Lebensmittelueberwachungsamt) complying with the EU directive 2010/63 and the german Animal Welfare Act §4(3) on the protection of animals used for scientific purposes.
Animals were bred and maintained for generations in-house in the institute’s fish facility (Leibniz-Institute on Aging - Fritz-Lipmann-institute, Beutenbergstr. 11, 07745 Jena, German) under animal husbandry license J-SHK-2684-04-08/11.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Baumgart M, Groth M, Priebe S, Savino A, Testa G, Dix A, Ripa R, Spallotta F, Gaetano C, Ori M, et al. RNA-Seq of the aging brain in the short-lived fish n. furzeri–conserved pathways and novel genes associated with neurogenesis. Aging Cell. 2014;13(6):965–974. doi: 10.1111/acel.12257. - DOI - PMC - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B (Methodol) 1995;57(1):289–300.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

