Protein recoding by ADAR1-mediated RNA editing is not essential for normal development and homeostasis
- PMID: 28874170
- PMCID: PMC5585977
- DOI: 10.1186/s13059-017-1301-4
Protein recoding by ADAR1-mediated RNA editing is not essential for normal development and homeostasis
Abstract
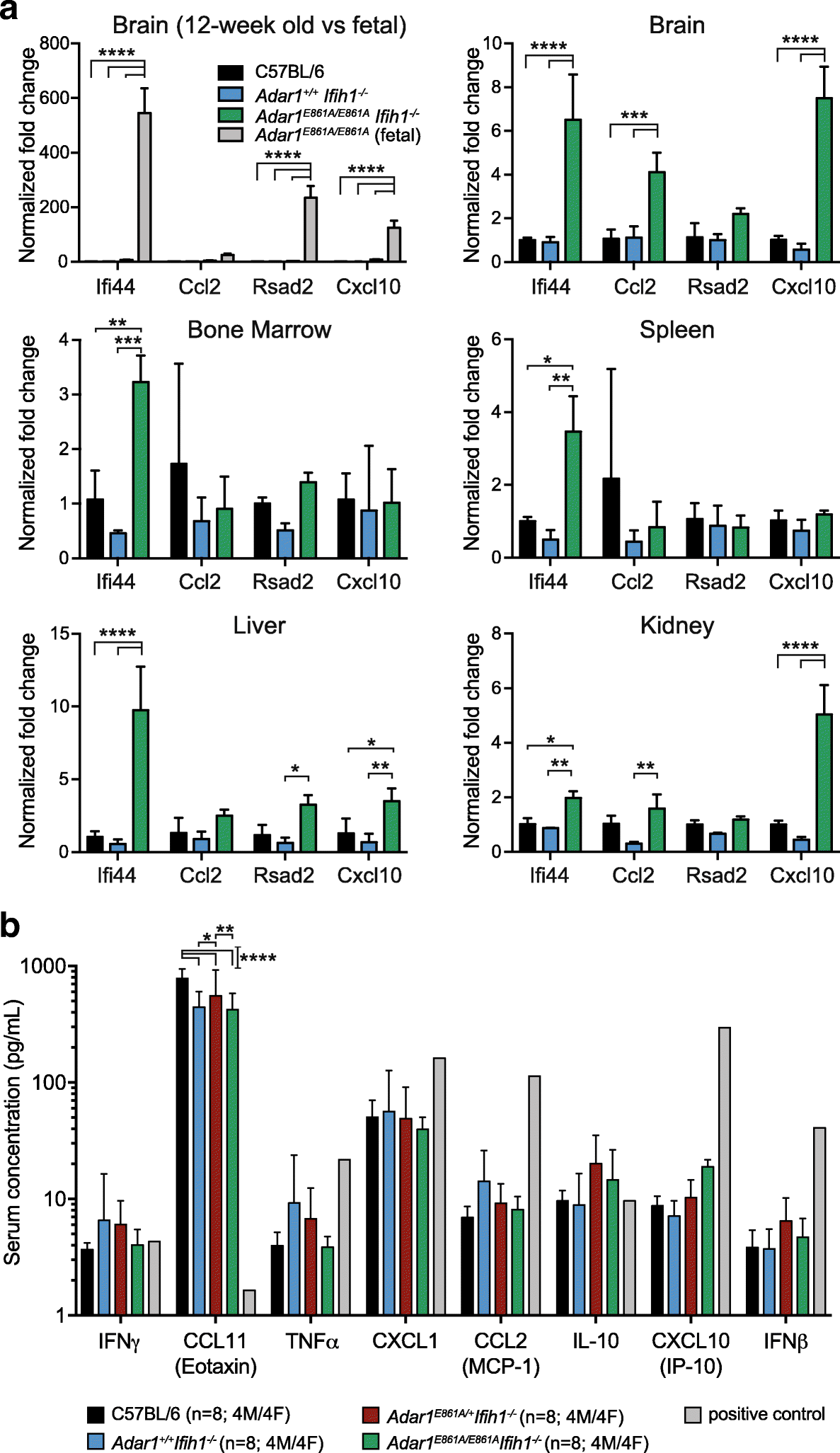
Background: Adenosine-to-inosine (A-to-I) editing of dsRNA by ADAR proteins is a pervasive epitranscriptome feature. Tens of thousands of A-to-I editing events are defined in the mouse, yet the functional impact of most is unknown. Editing causing protein recoding is the essential function of ADAR2, but an essential role for recoding by ADAR1 has not been demonstrated. ADAR1 has been proposed to have editing-dependent and editing-independent functions. The relative contribution of these in vivo has not been clearly defined. A critical function of ADAR1 is editing of endogenous RNA to prevent activation of the dsRNA sensor MDA5 (Ifih1). Outside of this, how ADAR1 editing contributes to normal development and homeostasis is uncertain.
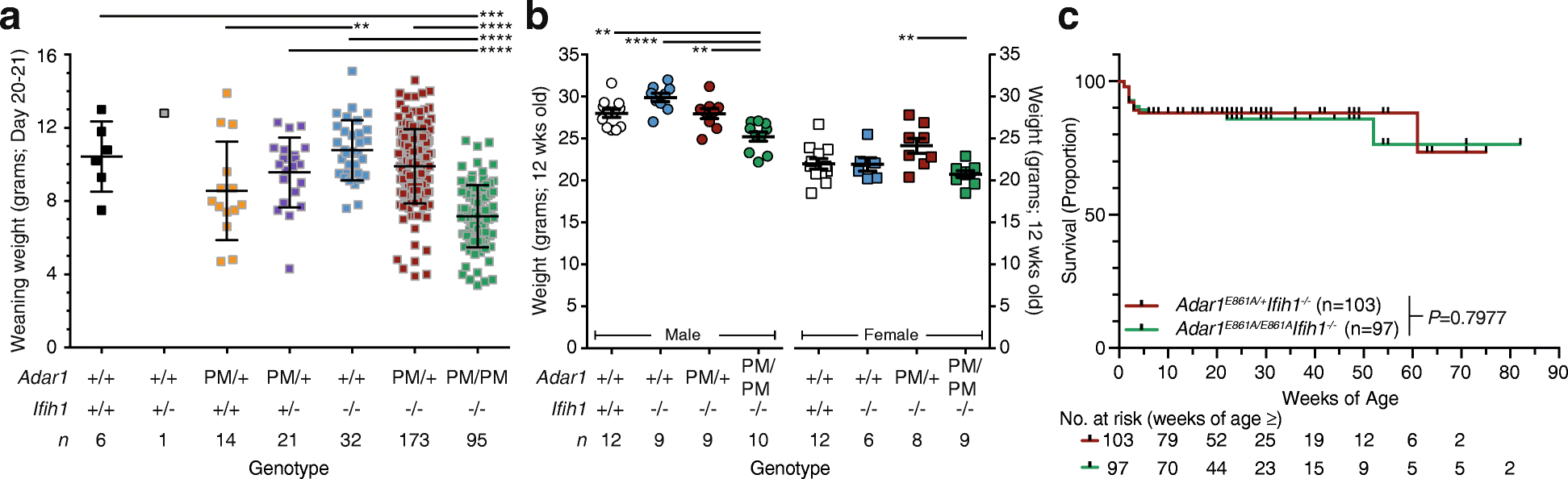
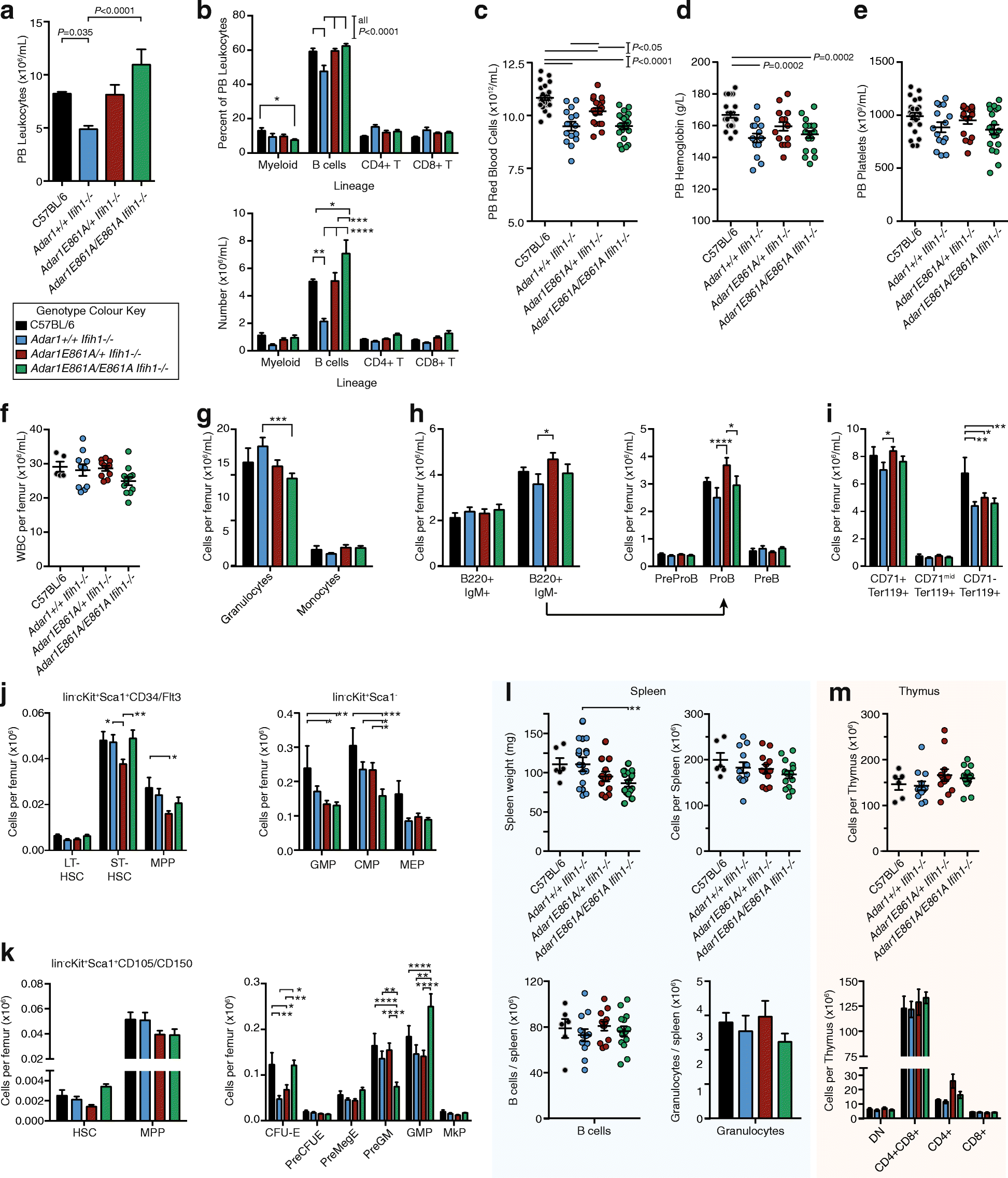
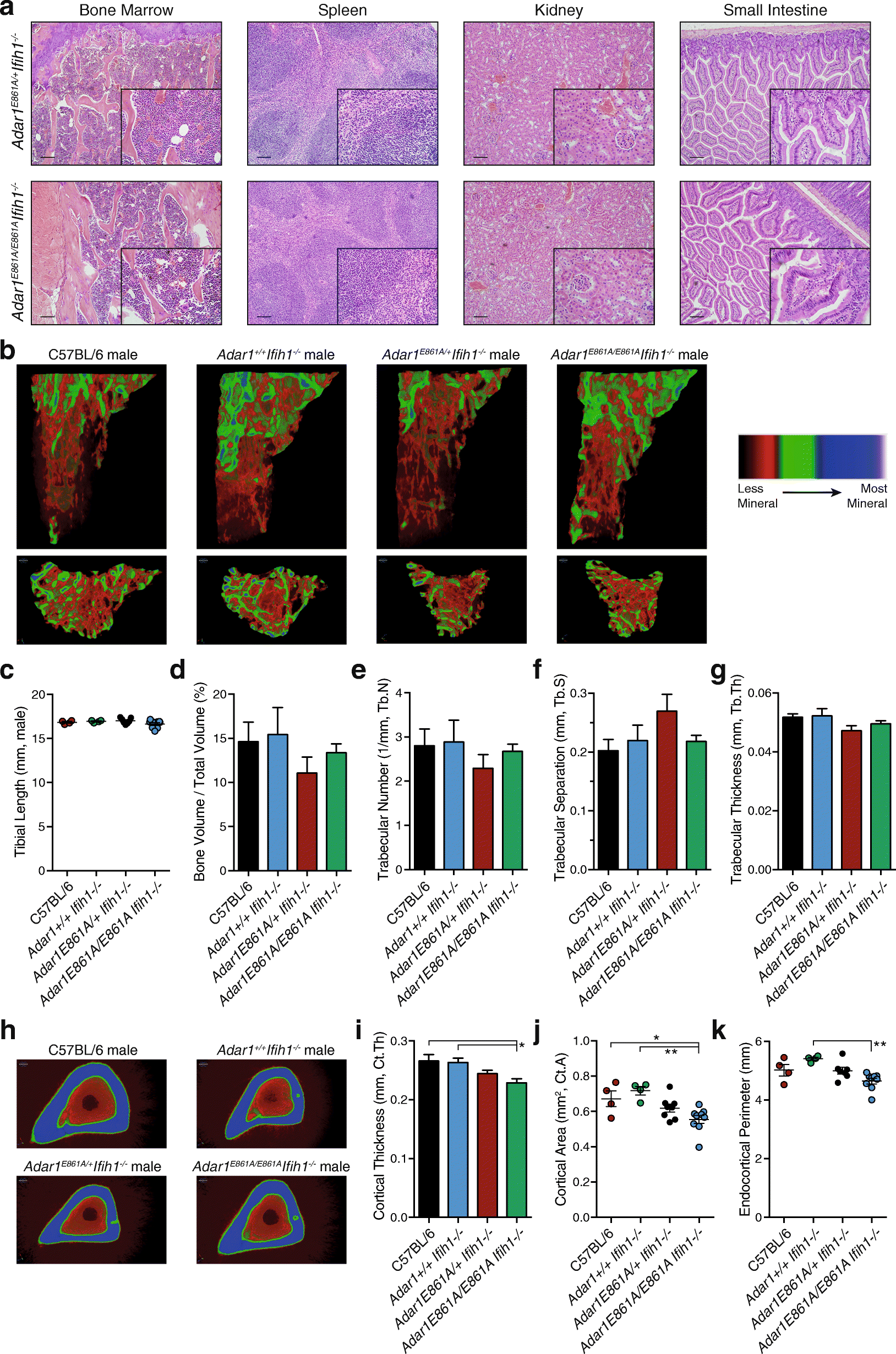
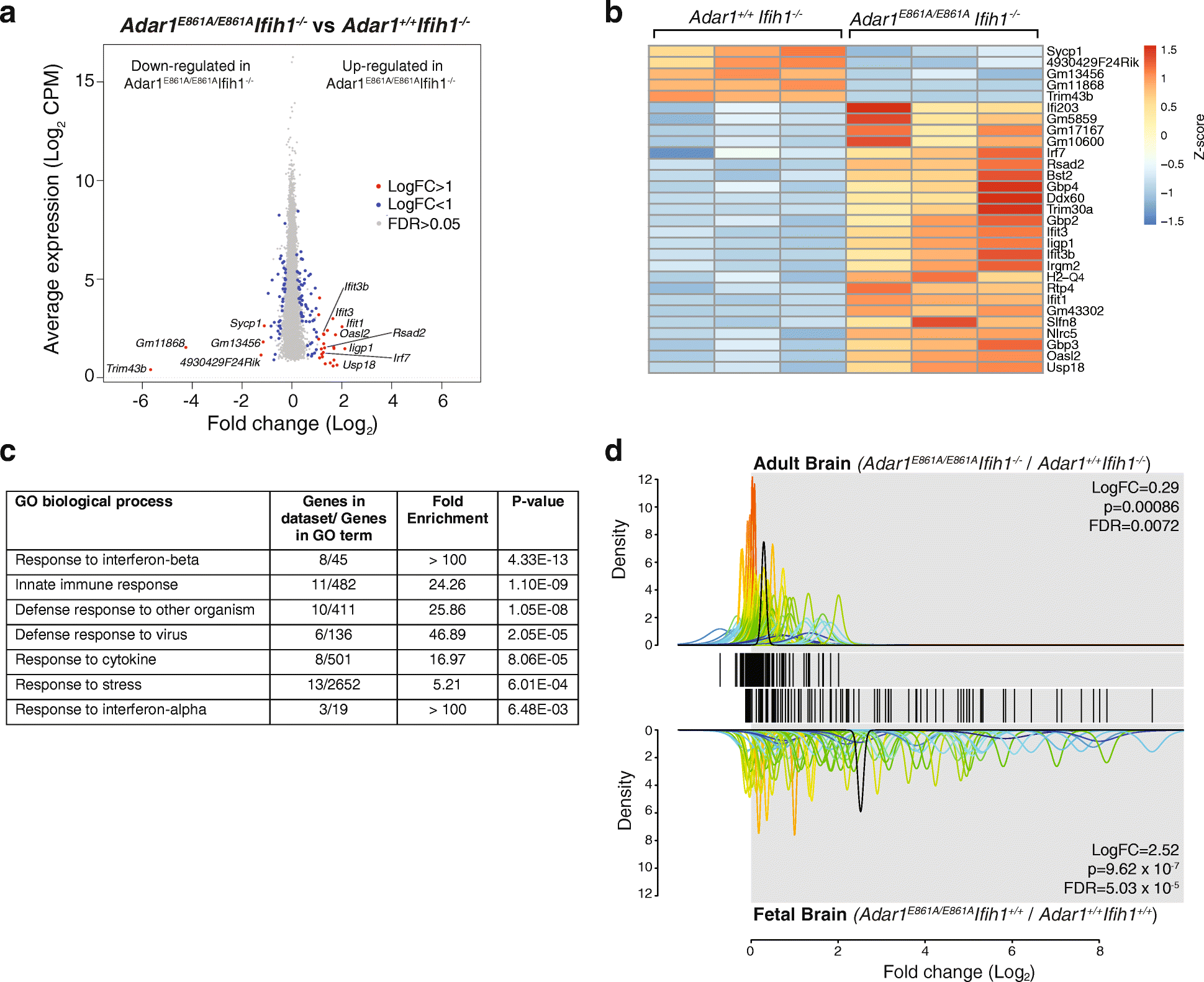
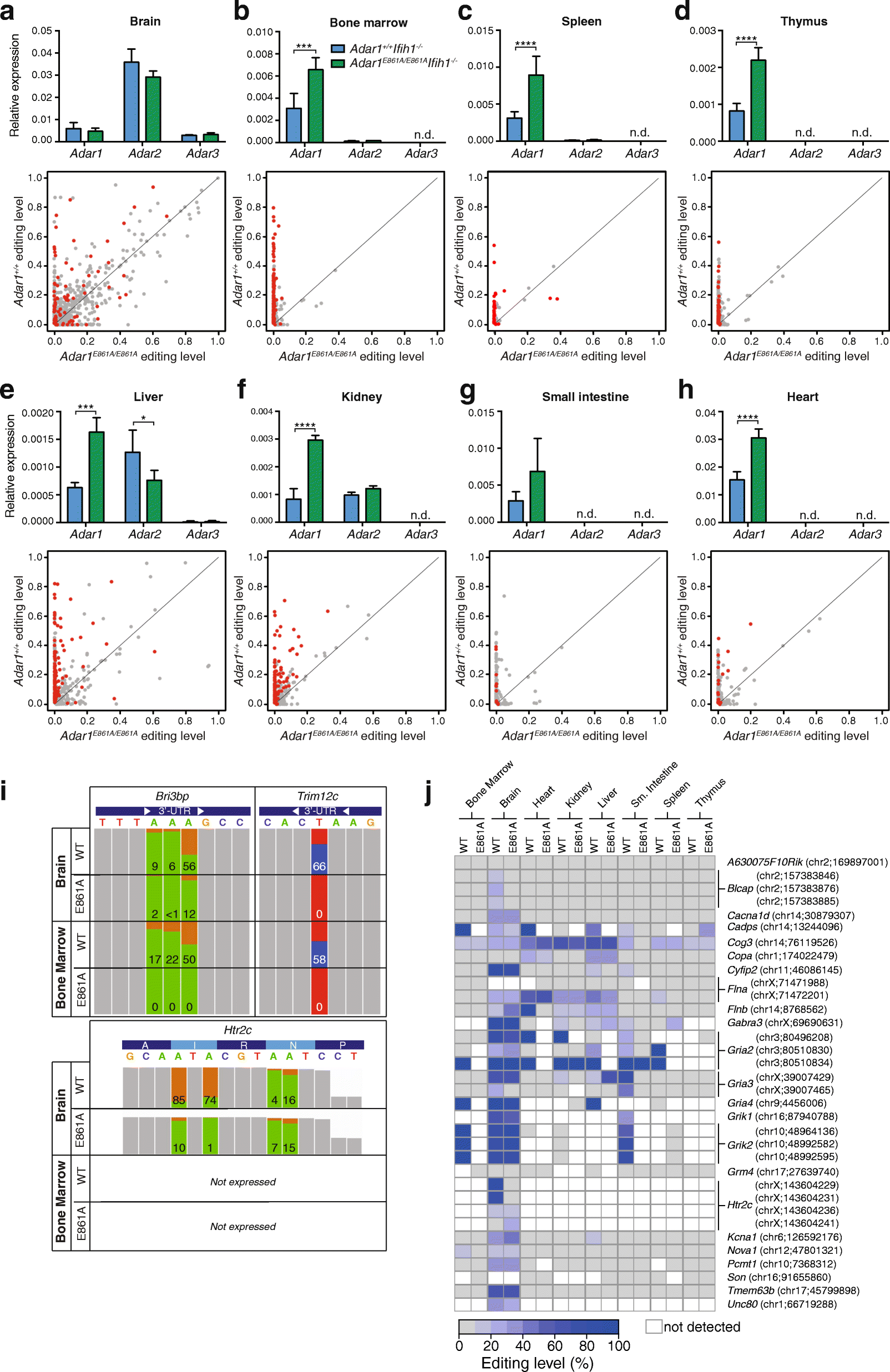
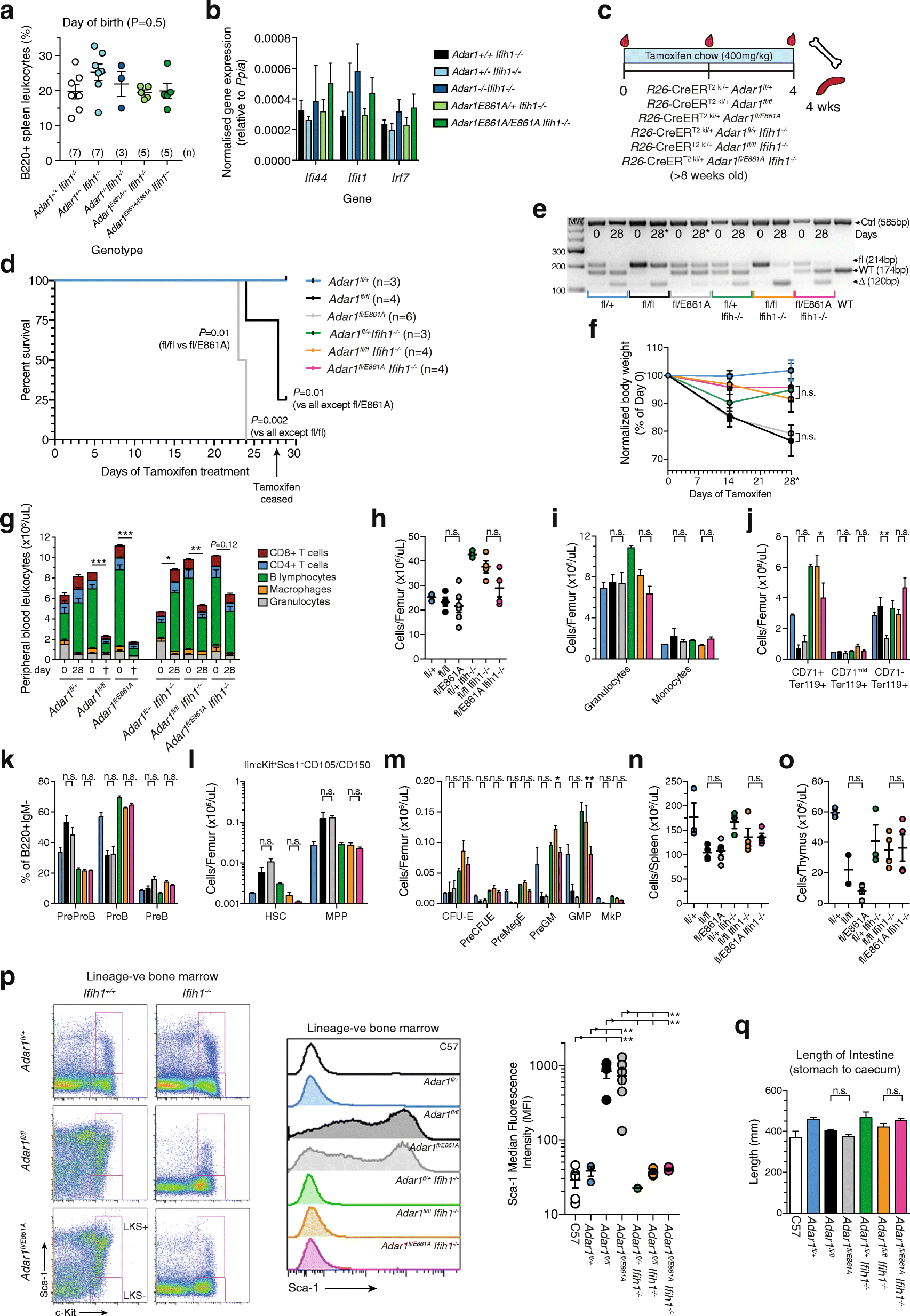
Results: We describe the consequences of ADAR1 editing deficiency on murine homeostasis. Adar1 E861A/E861A Ifih1 -/- mice are strikingly normal, including their lifespan. There is a mild, non-pathogenic innate immune activation signature in the Adar1 E861A/E861A Ifih1 -/- mice. Assessing A-to-I editing across adult tissues demonstrates that outside of the brain, ADAR1 performs the majority of editing and that ADAR2 cannot compensate in its absence. Direct comparison of the Adar1 -/- and Adar1 E861A/E861A alleles demonstrates a high degree of concordance on both Ifih1 +/+ and Ifih1 -/- backgrounds, suggesting no substantial contribution from ADAR1 editing-independent functions.
Conclusions: These analyses demonstrate that the lifetime absence of ADAR1-editing is well tolerated in the absence of MDA5. We conclude that protein recoding arising from ADAR1-mediated editing is not essential for organismal homeostasis. Additionally, the phenotypes associated with loss of ADAR1 are the result of RNA editing and MDA5-dependent functions.
Keywords: ADAR1; Development; Epitranscriptome; Innate immunity; MDA5; RNA editing; dsRNA.
Conflict of interest statement
Ethics approval and consent to participate
All animal experiments were approved by the St. Vincent’s Hospital, Melbourne Animal Ethics Committee (AEC#030/14 and AEC#031/15).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

