Genomics and Susceptibility Profiles of Extensively Drug-Resistant Pseudomonas aeruginosa Isolates from Spain
- PMID: 28874376
- PMCID: PMC5655108
- DOI: 10.1128/AAC.01589-17
Genomics and Susceptibility Profiles of Extensively Drug-Resistant Pseudomonas aeruginosa Isolates from Spain
Erratum in
-
Erratum for del Barrio-Tofiño et al., "Genomics and Susceptibility Profiles of Extensively Drug-Resistant Pseudomonas aeruginosa Isolates from Spain".Antimicrob Agents Chemother. 2017 Dec 21;62(1):e02352-17. doi: 10.1128/AAC.02352-17. Print 2018 Jan. Antimicrob Agents Chemother. 2017. PMID: 29269435 Free PMC article. No abstract available.
Abstract
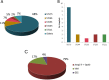
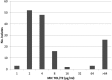
This study assessed the molecular epidemiology, resistance mechanisms, and susceptibility profiles of a collection of 150 extensively drug-resistant (XDR) Pseudomonas aeruginosa clinical isolates obtained from a 2015 Spanish multicenter study, with a particular focus on resistome analysis in relation to ceftolozane-tazobactam susceptibility. Broth microdilution MICs revealed that nearly all (>95%) of the isolates were nonsusceptible to piperacillin-tazobactam, ceftazidime, cefepime, aztreonam, imipenem, meropenem, and ciprofloxacin. Most of them were also resistant to tobramycin (77%), whereas nonsusceptibility rates were lower for ceftolozane-tazobactam (31%), amikacin (7%), and colistin (2%). Pulsed-field gel electrophoresis-multilocus sequence typing (PFGE-MLST) analysis revealed that nearly all of the isolates belonged to previously described high-risk clones. Sequence type 175 (ST175) was detected in all 9 participating hospitals and accounted for 68% (n = 101) of the XDR isolates, distantly followed by ST244 (n = 16), ST253 (n = 12), ST235 (n = 8), and ST111 (n = 2), which were detected only in 1 to 2 hospitals. Through phenotypic and molecular methods, the presence of horizontally acquired carbapenemases was detected in 21% of the isolates, mostly VIM (17%) and GES enzymes (4%). At least two representative isolates from each clone and hospital (n = 44) were fully sequenced on an Illumina MiSeq. Classical mutational mechanisms, such as those leading to the overexpression of the β-lactamase AmpC or efflux pumps, OprD inactivation, and/or quinolone resistance-determining regions (QRDR) mutations, were confirmed in most isolates and correlated well with the resistance phenotypes in the absence of horizontally acquired determinants. Ceftolozane-tazobactam resistance was not detected in carbapenemase-negative isolates, in agreement with sequencing data showing the absence of ampC mutations. The unique set of mutations responsible for the XDR phenotype of ST175 clone documented 7 years earlier were found to be conserved, denoting the long-term persistence of this specific XDR lineage in Spanish hospitals. Finally, other potentially relevant mutations were evidenced, including those in penicillin-binding protein 3 (PBP3), which is involved in β-lactam (including ceftolozane-tazobactam) resistance, and FusA1, which is linked to aminoglycoside resistance.
Keywords: Pseudomonas aeruginosa; extensively drug resistant; high-risk clones; whole-genome sequencing.
Copyright © 2017 American Society for Microbiology.
Figures

References
-
- Mesaros N, Nordmann P, Plésiat P, Roussel-Delvallez M, Van Eldere J, Glupczynski Y, Van Laethem Y, Jacobs F, Lebecque P, Malfroot A, Tulkens PM, Van Bambeke F. 2007. Pseudomonas aeruginosa: resistance and therapeutic options at the turn of the new millennium. Clin Microbiol Infect 13:560–578. doi: 10.1111/j.1469-0691.2007.01681.x. - DOI - PubMed
-
- Peña C, Suarez C, Gozalo M, Murillas J, Almirante B, Pomar V, Aguilar M, Granados A, Calbo E, Rodríguez-Baño J, Rodríguez F, Tubau F, Martínez-Martínez L, Oliver A, Spanish Network for Research in Infectious Diseases REIPI. 2012. Prospective multicenter study of the impact of carbapenem resistance on mortality in Pseudomonas aeruginosa bloodstream infections. Antimicrob Agents Chemother 56:1265–1272. doi: 10.1128/AAC.05991-11. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

