Mycobacterium abscessus WhiB7 Regulates a Species-Specific Repertoire of Genes To Confer Extreme Antibiotic Resistance
- PMID: 28874378
- PMCID: PMC5655061
- DOI: 10.1128/AAC.01347-17
Mycobacterium abscessus WhiB7 Regulates a Species-Specific Repertoire of Genes To Confer Extreme Antibiotic Resistance
Abstract
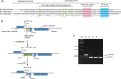
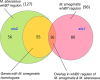
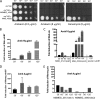
Mycobacterium abscessus causes acute and chronic bronchopulmonary infection in patients with chronic lung damage, of which cystic fibrosis (CF) patients are particularly vulnerable. The major threat posed by this organism is its high intrinsic antibiotic resistance. A typical treatment regimen involves a 6- to 12-month-long combination therapy of clarithromycin and amikacin, with cure rates below 50% and multiple side effects, especially due to amikacin. In the present work, we show that M. abscessuswhiB7, a homologue of Mycobacterium tuberculosis and Mycobacterium smegmatis whiB7 with previously demonstrated effects on intrinsic antibiotic resistance, is strongly induced when exposed to clinically relevant antibiotics that target the ribosome: erythromycin, clarithromycin, amikacin, tetracycline, and spectinomycin. The deletion of M. abscessuswhiB7 results in sensitivity to all of the above-mentioned antibiotics. Further, we have defined and compared the whiB7 regulon of M. abscessus with the closely related nontuberculous mycobacterium (NTM) M. smegmatis to demonstrate the induction of a species-specific repertoire of genes. Finally, we show that one such gene, eis2, is specifically induced in M. abscessus by whiB7 and contributes to its higher levels of intrinsic amikacin resistance. This species-specific pattern of gene induction might account for the differences in drug susceptibilities to other antibiotics and between different mycobacterial species.
Keywords: Mycobacterium; abscessus; antibiotic resistance; intrinsic; whiB7.
Copyright © 2017 American Society for Microbiology.
Figures



References
-
- Prevots DR, Shaw PA, Strickland D, Jackson LA, Raebel MA, Blosky MA, Montes de Oca R, Shea YR, Seitz AE, Holland SM, Olivier KN. 2010. Nontuberculous mycobacterial lung disease prevalence at four integrated health care delivery systems. Am J Respir Crit Care Med 182:970–976. doi: 10.1164/rccm.201002-0310OC. - DOI - PMC - PubMed
-
- Qvist T, Gilljam M, Jonsson B, Taylor-Robinson D, Jensen-Fangel S, Wang M, Svahn A, Kotz K, Hansson L, Hollsing A, Hansen CR, Finstad PL, Pressler T, Hoiby N, Katzenstein TL, Scandinavian Cystic Fibrosis Study Consortium (SCFSC). 2015. Epidemiology of nontuberculous mycobacteria among patients with cystic fibrosis in Scandinavia. J Cyst Fibros 14:46–52. doi: 10.1016/j.jcf.2014.08.002. - DOI - PMC - PubMed
-
- Olivier KN, Weber DJ, Wallace RJ Jr, Faiz AR, Lee JH, Zhang Y, Brown-Elliot BA, Handler A, Wilson RW, Schechter MS, Edwards LJ, Chakraborti S, Knowles MR, Nontuberculous Mycobacteria in Cystic Fibrosis Study Group. 2003. Nontuberculous mycobacteria. I: multicenter prevalence study in cystic fibrosis. Am J Respir Crit Care Med 167:828–834. doi: 10.1164/rccm.200207-678OC. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

