Use of next-generation sequencing to detect LDLR gene copy number variation in familial hypercholesterolemia
- PMID: 28874442
- PMCID: PMC5665663
- DOI: 10.1194/jlr.D079301
Use of next-generation sequencing to detect LDLR gene copy number variation in familial hypercholesterolemia
Abstract
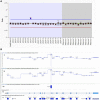
Familial hypercholesterolemia (FH) is a heritable condition of severely elevated LDL cholesterol, caused predominantly by autosomal codominant mutations in the LDL receptor gene (LDLR). In providing a molecular diagnosis for FH, the current procedure often includes targeted next-generation sequencing (NGS) panels for the detection of small-scale DNA variants, followed by multiplex ligation-dependent probe amplification (MLPA) in LDLR for the detection of whole-exon copy number variants (CNVs). The latter is essential because ∼10% of FH cases are attributed to CNVs in LDLR; accounting for them decreases false negative findings. Here, we determined the potential of replacing MLPA with bioinformatic analysis applied to NGS data, which uses depth-of-coverage analysis as its principal method to identify whole-exon CNV events. In analysis of 388 FH patient samples, there was 100% concordance in LDLR CNV detection between these two methods: 38 reported CNVs identified by MLPA were also successfully detected by our NGS method, while 350 samples negative for CNVs by MLPA were also negative by NGS. This result suggests that MLPA can be removed from the routine diagnostic screening for FH, significantly reducing associated costs, resources, and analysis time, while promoting more widespread assessment of this important class of mutations across diagnostic laboratories.
Keywords: DNA variation; LDL; bioinformatics; coronary heart disease; diagnostic tools; genetic testing; lipid and lipoprotein metabolism; lipoprotein receptors; molecular biology/genetics; precision medicine.
Copyright © 2017 by the American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
R.A.H. has received honoraria for membership on advisory boards and speakers’ bureaus for Aegerion, Amgen, Gemphire, Eli Lilly and Company, Merck, Pfizer, Regeneron Pharmaceuticals, Sanofi, and Valeant Pharmaceuticals International. M.A.I. has no disclosures.
Figures

References
-
- Nordestgaard B. G., Chapman M. J., Humphries S. E., Ginsberg H. N., Masana L., Descamps O. S., Wiklund O., Hegele R. A., Raal F. J., Defesche J. C., et al. . 2013. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European atherosclerosis society. Eur. Heart J. 34: 3478–3490. - PMC - PubMed
-
- Usifo E., Leigh S. E. A., Whittall R. A., Lench N., Taylor A., Yeats C., Orengo C. A., Martin A. C. R., Celli J., and Humphries S. E.. 2012. Low-density lipoprotein receptor gene familial hypercholesterolemia variant database: update and pathological assessment. Ann. Hum. Genet. 76: 387–401. - PubMed
-
- Pollex R. L., and Hegele R. A.. 2007. Copy number variation in the human genome and its implications for cardiovascular disease. Circulation. 115: 3130–3138. - PubMed
-
- Wald D. S., Bestwick J. P., Morris J. K., Whyte K., Jenkins L., and Wald N. J.. 2016. Child-parent familial hypercholesterolemia screening in primary care. N. Engl. J. Med. 375: 1628–1637. - PubMed
-
- Sjouke B., Kusters D. M., Kindt I., Besseling J., Defesche J. C., Sijbrands E. J. G., Roeters van Lennep J. E., Stalenhoef A. F. H., Wiegman A., de Graaf J., et al. . 2015. Homozygous autosomal dominant hypercholesterolaemia in the Netherlands: prevalence, genotype-phenotype relationship, and clinical outcome. Eur. Heart J. 36: 560–565. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

