Internal Associations of the Acidic Region of Upstream Binding Factor Control Its Nucleolar Localization
- PMID: 28874518
- PMCID: PMC5660467
- DOI: 10.1128/MCB.00218-17
Internal Associations of the Acidic Region of Upstream Binding Factor Control Its Nucleolar Localization
Abstract
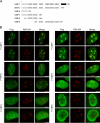
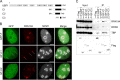
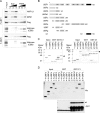
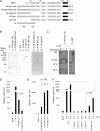
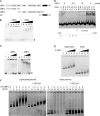
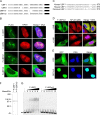
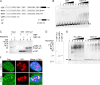
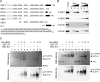
Upstream binding factor (UBF) is a member of the high-mobility group (HMG) box protein family, characterized by multiple HMG boxes and a C-terminal acidic region (AR). UBF is an essential transcription factor for rRNA genes and mediates the formation of transcriptionally active chromatin in the nucleolus. However, it remains unknown how UBF is specifically localized to the nucleolus. Here, we examined the molecular mechanisms that localize UBF to the nucleolus. We found that the first HMG box (HMG box 1), the linker region (LR), and the AR cooperatively regulate the nucleolar localization of UBF1. We demonstrated that the AR intramolecularly associates with and attenuates the DNA binding activity of HMG boxes and confers the structured DNA preference to HMG box 1. In contrast, the LR was found to serve as a nuclear localization signal and compete with HMG boxes to bind the AR, permitting nucleolar localization of UBF1. The LR sequence binds DNA and assists the stable chromatin binding of UBF. We also showed that the phosphorylation status of the AR does not clearly affect the localization of UBF1. Our results strongly suggest that associations of the AR with HMG boxes and the LR regulate UBF nucleolar localization.
Keywords: HMG box; nucleolus; ribosome biogenesis; transcription factors.
Copyright © 2017 American Society for Microbiology.
Figures
References
-
- Roussel P, Andre C, Masson C, Geraud G, Hernandez-Verdun D. 1993. Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J Cell Sci 104(Part 2):327–337. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
