Identification of individuals by trait prediction using whole-genome sequencing data
- PMID: 28874526
- PMCID: PMC5617305
- DOI: 10.1073/pnas.1711125114
Identification of individuals by trait prediction using whole-genome sequencing data
Erratum in
-
Correction for Lippert et al., Identification of individuals by trait prediction using whole-genome sequencing data.Proc Natl Acad Sci U S A. 2017 Oct 10;114(41):E8800. doi: 10.1073/pnas.1716166114. Epub 2017 Oct 2. Proc Natl Acad Sci U S A. 2017. PMID: 28973936 Free PMC article. No abstract available.
Abstract
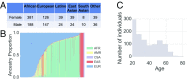
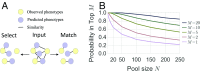
Prediction of human physical traits and demographic information from genomic data challenges privacy and data deidentification in personalized medicine. To explore the current capabilities of phenotype-based genomic identification, we applied whole-genome sequencing, detailed phenotyping, and statistical modeling to predict biometric traits in a cohort of 1,061 participants of diverse ancestry. Individually, for a large fraction of the traits, their predictive accuracy beyond ancestry and demographic information is limited. However, we have developed a maximum entropy algorithm that integrates multiple predictions to determine which genomic samples and phenotype measurements originate from the same person. Using this algorithm, we have reidentified an average of >8 of 10 held-out individuals in an ethnically mixed cohort and an average of 5 of either 10 African Americans or 10 Europeans. This work challenges current conceptions of personal privacy and may have far-reaching ethical and legal implications.
Keywords: DNA phenotyping; genome sequencing; genomic privacy; phenotype prediction; reidentification.
Conflict of interest statement
Conflict of interest statement: The authors are employees of and own equity in Human Longevity Inc.
Figures



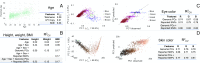

Comment in
-
Geneticists pan paper that claims to predict a person's face from their DNA.Nature. 2017 Sep 8;549(7671):139-140. doi: 10.1038/nature.2017.22580. Nature. 2017. PMID: 28905923 No abstract available.
References
-
- Frudakis T. Molecular Photofitting: Predicting Ancestry and Phenotype Using DNA. Elsevier; New York: 2010.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

