Effects of thymic selection on T cell recognition of foreign and tumor antigenic peptides
- PMID: 28874554
- PMCID: PMC5617294
- DOI: 10.1073/pnas.1708573114
Effects of thymic selection on T cell recognition of foreign and tumor antigenic peptides
Abstract
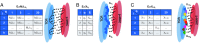
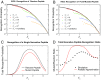
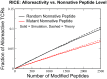
The advent of cancer immunotherapy has generated renewed hope for the treatment of many malignancies by introducing a number of novel strategies that exploit various properties of the immune system. These therapies are based on the idea that cytotoxic T lymphocytes (CTLs) directly recognize and respond to tumor-associated neoantigens (TANs) in much the same way as they would to foreign peptides presented on cell surfaces. To date, however, nearly all attempts to optimize immunotherapeutic strategies have been empirical. Here, we develop a model of T cell selection based on the assumption of random interaction strengths between a self-peptide and the various T cell receptors. The model enables the analytical study of the effects of selection on the CTL recognition of TANs and completely foreign peptides and can estimate the number of CTLs that can detect donor-matched transplants. We show that negative selection thresholds chosen to reflect experimentally observed thymic survival rates result in near-optimal production of T cells that are capable of surviving selection and recognizing foreign antigen. These analytical results are confirmed by simulation.
Keywords: T cell; applied probability; immunotherapy; neoantigen.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The present status and future prospects of peptide-based cancer vaccines.Int Immunol. 2016 Jul;28(7):319-28. doi: 10.1093/intimm/dxw027. Epub 2016 May 28. Int Immunol. 2016. PMID: 27235694 Review.
-
Different affinity windows for virus and cancer-specific T-cell receptors: implications for therapeutic strategies.Eur J Immunol. 2012 Dec;42(12):3174-9. doi: 10.1002/eji.201242606. Epub 2012 Oct 16. Eur J Immunol. 2012. PMID: 22949370 Free PMC article.
-
Harnessing the power of Vδ2 cells in cancer immunotherapy.Clin Exp Immunol. 2015 Apr;180(1):1-10. doi: 10.1111/cei.12564. Clin Exp Immunol. 2015. PMID: 25469879 Free PMC article. Review.
-
Development of lymphopoiesis as a function of the thymic microenvironment. Use of CD8+ cytotoxic T lymphocytes for cellular immunotherapy of human cancer.In Vivo. 1994 Nov-Dec;8(5):915-43. In Vivo. 1994. PMID: 7727741 Review.
-
The role of neoantigens in response to immune checkpoint blockade.Int Immunol. 2016 Aug;28(8):411-9. doi: 10.1093/intimm/dxw019. Epub 2016 Apr 5. Int Immunol. 2016. PMID: 27048318 Free PMC article. Review.
Cited by
-
A Proposed Link Between Acute Thymic Involution and Late Adverse Effects of Chemotherapy.Front Immunol. 2022 Jul 1;13:933547. doi: 10.3389/fimmu.2022.933547. eCollection 2022. Front Immunol. 2022. PMID: 35844592 Free PMC article.
-
Rapid Assessment of T-Cell Receptor Specificity of the Immune Repertoire.Nat Comput Sci. 2021 May;1(5):362-373. doi: 10.1038/s43588-021-00076-1. Epub 2021 May 24. Nat Comput Sci. 2021. PMID: 36090450 Free PMC article.
-
Immune cells use active tugging forces to distinguish affinity and accelerate evolution.Proc Natl Acad Sci U S A. 2023 Mar 14;120(11):e2213067120. doi: 10.1073/pnas.2213067120. Epub 2023 Mar 10. Proc Natl Acad Sci U S A. 2023. PMID: 36897986 Free PMC article.
-
RACER-m leverages structural features for sparse T cell specificity prediction.Sci Adv. 2024 May 17;10(20):eadl0161. doi: 10.1126/sciadv.adl0161. Epub 2024 May 15. Sci Adv. 2024. PMID: 38748791 Free PMC article.
-
How persistent infection overcomes peripheral tolerance mechanisms to cause T cell-mediated autoimmune disease.Proc Natl Acad Sci U S A. 2024 Mar 12;121(11):e2318599121. doi: 10.1073/pnas.2318599121. Epub 2024 Mar 6. Proc Natl Acad Sci U S A. 2024. PMID: 38446856 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

