A mutation in the melon Vacuolar Protein Sorting 41prevents systemic infection of Cucumber mosaic virus
- PMID: 28874719
- PMCID: PMC5585375
- DOI: 10.1038/s41598-017-10783-3
A mutation in the melon Vacuolar Protein Sorting 41prevents systemic infection of Cucumber mosaic virus
Abstract
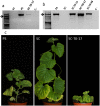
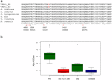
In the melon exotic accession PI 161375, the gene cmv1, confers recessive resistance to Cucumber mosaic virus (CMV) strains of subgroup II. cmv1 prevents the systemic infection by restricting the virus to the bundle sheath cells and impeding viral loading to the phloem. Here we report the fine mapping and cloning of cmv1. Screening of an F2 population reduced the cmv1 region to a 132 Kb interval that includes a Vacuolar Protein Sorting 41 gene. CmVPS41 is conserved among plants, animals and yeast and is required for post-Golgi vesicle trafficking towards the vacuole. We have validated CmVPS41 as the gene responsible for the resistance, both by generating CMV susceptible transgenic melon plants, expressing the susceptible allele in the resistant cultivar and by characterizing CmVPS41 TILLING mutants with reduced susceptibility to CMV. Finally, a core collection of 52 melon accessions allowed us to identify a single amino acid substitution (L348R) as the only polymorphism associated with the resistant phenotype. CmVPS41 is the first natural recessive resistance gene found to be involved in viral transport and its cellular function suggests that CMV might use CmVPS41 for its own transport towards the phloem.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

