Protein crystallography and drug discovery: recollections of knowledge exchange between academia and industry
- PMID: 28875019
- PMCID: PMC5571795
- DOI: 10.1107/S2052252517009241
Protein crystallography and drug discovery: recollections of knowledge exchange between academia and industry
Abstract
The development of structure-guided drug discovery is a story of knowledge exchange where new ideas originate from all parts of the research ecosystem. Dorothy Crowfoot Hodgkin obtained insulin from Boots Pure Drug Company in the 1930s and insulin crystallization was optimized in the company Novo in the 1950s, allowing the structure to be determined at Oxford University. The structure of renin was developed in academia, on this occasion in London, in response to a need to develop antihypertensives in pharma. The idea of a dimeric aspartic protease came from an international academic team and was discovered in HIV; it eventually led to new HIV antivirals being developed in industry. Structure-guided fragment-based discovery was developed in large pharma and biotechs, but has been exploited in academia for the development of new inhibitors targeting protein-protein interactions and also antimicrobials to combat mycobacterial infections such as tuberculosis. These observations provide a strong argument against the so-called 'linear model', where ideas flow only in one direction from academic institutions to industry. Structure-guided drug discovery is a story of applications of protein crystallography and knowledge exhange between academia and industry that has led to new drug approvals for cancer and other common medical conditions by the Food and Drug Administration in the USA, as well as hope for the treatment of rare genetic diseases and infectious diseases that are a particular challenge in the developing world.
Keywords: cancer; disease; fragment-based structure-guided drug discovery; protein crystallography; protein structure.
Figures


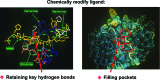
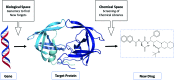

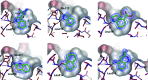

References
-
- Adams, M. J., Blundell, T. L., Dodson, E. J., Dodson, G. G., Vijayan, M., Baker, E. N., Harding, M. M., Hodgkin, D. C., Rimmer, B. & Sheat, S. (1969). Nature (London), 224, 491–495.
-
- Andreeva, N. S., Zdanov, A. S., Gustchina, A. E. & Fedorov, A. A. (1984). J. Biol. Chem. 259, 11353–11365. - PubMed
-
- Arnold, E. (2014). Prog. Biophys. Mol. Biol. 116, 81. - PubMed
-
- Atkinson, A. B. (1980). The Therapeutics of Hypertension, edited by J. I. S. Robertson, G. W. Pickering & A. D. S. Caldwell, pp. 29–61. London: The Royal Society of Medicine.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

