Silencing CAFFEOYL SHIKIMATE ESTERASE Affects Lignification and Improves Saccharification in Poplar
- PMID: 28878037
- PMCID: PMC5664470
- DOI: 10.1104/pp.17.00920
Silencing CAFFEOYL SHIKIMATE ESTERASE Affects Lignification and Improves Saccharification in Poplar
Abstract
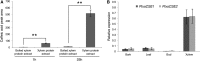
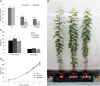
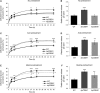
Caffeoyl shikimate esterase (CSE) was recently shown to play an essential role in lignin biosynthesis in Arabidopsis (Arabidopsis thaliana) and later in Medicago truncatula However, the general function of this enzyme was recently questioned by the apparent lack of CSE activity in lignifying tissues of different plant species. Here, we show that down-regulation of CSE in hybrid poplar (Populus tremula × Populus alba) resulted in up to 25% reduced lignin deposition, increased levels of p-hydroxyphenyl units in the lignin polymer, and a relatively higher cellulose content. The transgenic trees were morphologically indistinguishable from the wild type. Ultra-high-performance liquid chromatography-mass spectrometry-based phenolic profiling revealed a reduced abundance of several oligolignols containing guaiacyl and syringyl units and their corresponding hydroxycinnamaldehyde units, in agreement with the reduced flux toward coniferyl and sinapyl alcohol. These trees accumulated the CSE substrate caffeoyl shikimate along with other compounds belonging to the metabolic classes of benzenoids and hydroxycinnamates. Furthermore, the reduced lignin amount combined with the relative increase in cellulose content in the CSE down-regulated lines resulted in up to 62% more glucose released per plant upon limited saccharification when no pretreatment was applied and by up to 86% and 91% when acid and alkaline pretreatments were used. Our results show that CSE is not only important for the lignification process in poplar but is also a promising target for the development of improved lignocellulosic biomass crops for sugar platform biorefineries.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures



References
-
- Berlin A, Balakshin M, Gilkes N, Kadla J, Maximenko V, Kubo S, Saddler J (2006) Inhibition of cellulase, xylanase and β-glucosidase activities by softwood lignin preparations. J Biotechnol 125: 198–209 - PubMed
-
- Bjurhager I, Olsson AM, Zhang B, Gerber L, Kumar M, Berglund LA, Burgert I, Sundberg B, Salmén L (2010) Ultrastructure and mechanical properties of Populus wood with reduced lignin content caused by transgenic down-regulation of cinnamate 4-hydroxylase. Biomacromolecules 11: 2359–2365 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

