Lysine 242 within Helix 10 of the Pseudorabies Virus Nuclear Egress Complex pUL31 Component Is Critical for Primary Envelopment of Nucleocapsids
- PMID: 28878082
- PMCID: PMC5660471
- DOI: 10.1128/JVI.01182-17
Lysine 242 within Helix 10 of the Pseudorabies Virus Nuclear Egress Complex pUL31 Component Is Critical for Primary Envelopment of Nucleocapsids
Abstract
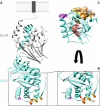
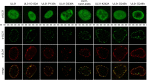
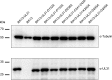
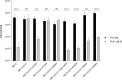
Newly assembled herpesvirus nucleocapsids are translocated from the nucleus to the cytosol by a vesicle-mediated process engaging the nuclear membranes. This transport is governed by the conserved nuclear egress complex (NEC), consisting of the alphaherpesviral pUL34 and pUL31 homologs. The NEC is not only required for efficient nuclear egress but also sufficient for vesicle formation from the inner nuclear membrane (INM), as well as from synthetic lipid bilayers. The recently solved crystal structures for the NECs from different herpesviruses revealed molecular details of this membrane deformation and scission machinery uncovering the interfaces involved in complex and coat formation. However, the interaction domain with the nucleocapsid remained undefined. Since the NEC assembles a curved hexagonal coat on the nucleoplasmic side of the INM consisting of tightly interwoven pUL31/pUL34 heterodimers arranged in hexamers, only the membrane-distal end of the NEC formed by pUL31 residues appears to be accessible for interaction with the nucleocapsid cargo. To identify the amino acids involved in capsid incorporation, we mutated the corresponding regions in the alphaherpesvirus pseudorabies virus (PrV). Site-specifically mutated pUL31 homologs were tested for localization, interaction with pUL34, and complementation of PrV-ΔUL31. We identified a conserved lysine residue at amino acid position 242 in PrV pUL31 located in the alpha-helical domain H10 exposed on the membrane-distal end of the NEC as a key residue for nucleocapsid incorporation into the nascent primary particle.IMPORTANCE Vesicular transport through the nuclear envelope is a focus of research but is still not well understood. Herpesviruses pioneered this mechanism for translocation of the newly assembled nucleocapsid from the nucleus into the cytosol via vesicles derived from the inner nuclear membrane which fuse in a well-tuned process with the outer nuclear membrane to release their content. The structure of the viral nuclear membrane budding and scission machinery has been solved recently, providing in-depth molecular details. However, how cargo is incorporated remained unclear. We identified a conserved lysine residue in the membrane-distal portion of the nuclear egress complex required for capsid uptake into inner nuclear membrane-derived vesicles.
Keywords: NEC capsid interaction; herpesvirus; nuclear egress complex (NEC); nucleocapsid; pUL31; pUL34; pseudorabies virus (PrV).
Copyright © 2017 American Society for Microbiology.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

