Exome-capture RNA sequencing of decade-old breast cancers and matched decalcified bone metastases
- PMID: 28878133
- PMCID: PMC5621874
- DOI: 10.1172/jci.insight.95703
Exome-capture RNA sequencing of decade-old breast cancers and matched decalcified bone metastases
Abstract
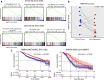
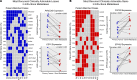
Bone metastases (BoM) are a significant cause of morbidity in patients with estrogen receptor-positive (ER-positive) breast cancer; yet, characterizations of human specimens are limited. In this study, exome-capture RNA sequencing (ecRNA-seq) on aged (8-12 years), formalin-fixed, paraffin-embedded (FFPE), and decalcified cancer specimens was evaluated. Gene expression values and ecRNA-seq quality metrics from FFPE or decalcified tumor RNA showed minimal differences when compared with matched flash-frozen or nondecalcified tumors. ecRNA-seq was then applied on a longitudinal collection of 11 primary breast cancers and patient-matched synchronous or recurrent BoMs. Overtime, BoMs exhibited gene expression shifts to more Her2 and LumB PAM50 subtype profiles, temporally influenced expression evolution, recurrently dysregulated prognostic gene sets, and longitudinal expression alterations of clinically actionable genes, particularly in the CDK/Rb/E2F and FGFR signaling pathways. Taken together, this study demonstrates the use of ecRNA-seq on decade-old and decalcified specimens and defines recurrent longitudinal transcriptional remodeling events in estrogen-deprived breast cancers.
Keywords: Breast cancer; Genetics; Oncology.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

