Reconstructing cell cycle and disease progression using deep learning
- PMID: 28878212
- PMCID: PMC5587733
- DOI: 10.1038/s41467-017-00623-3
Reconstructing cell cycle and disease progression using deep learning
Abstract
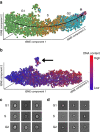
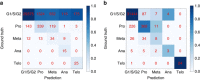
We show that deep convolutional neural networks combined with nonlinear dimension reduction enable reconstructing biological processes based on raw image data. We demonstrate this by reconstructing the cell cycle of Jurkat cells and disease progression in diabetic retinopathy. In further analysis of Jurkat cells, we detect and separate a subpopulation of dead cells in an unsupervised manner and, in classifying discrete cell cycle stages, we reach a sixfold reduction in error rate compared to a recent approach based on boosting on image features. In contrast to previous methods, deep learning based predictions are fast enough for on-the-fly analysis in an imaging flow cytometer.The interpretation of information-rich, high-throughput single-cell data is a challenge requiring sophisticated computational tools. Here the authors demonstrate a deep convolutional neural network that can classify cell cycle status on-the-fly.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Brown M, Wittwer C. Flow cytometry: principles and clinical applications in hematology. Clin. Chem. 2000;46:1221–1229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

