APOBEC3A is an oral cancer prognostic biomarker in Taiwanese carriers of an APOBEC deletion polymorphism
- PMID: 28878238
- PMCID: PMC5587710
- DOI: 10.1038/s41467-017-00493-9
APOBEC3A is an oral cancer prognostic biomarker in Taiwanese carriers of an APOBEC deletion polymorphism
Abstract
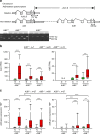
Oral squamous cell carcinoma is a prominent cancer worldwide, particularly in Taiwan. By integrating omics analyses in 50 matched samples, we uncover in Taiwanese patients a predominant mutation signature associated with cytidine deaminase APOBEC, which correlates with the upregulation of APOBEC3A expression in the APOBEC3 gene cluster at 22q13. APOBEC3A expression is significantly higher in tumors carrying APOBEC3B-deletion allele(s). High-level APOBEC3A expression is associated with better overall survival, especially among patients carrying APOBEC3B-deletion alleles, as examined in a second cohort (n = 188; p = 0.004). The frequency of APOBEC3B-deletion alleles is ~50% in 143 genotyped oral squamous cell carcinoma -Taiwan samples (27A3B -/-:89A3B +/-:27A3B +/+), compared to the 5.8% found in 314 OSCC-TCGA samples. We thus report a frequent APOBEC mutational profile, which relates to a APOBEC3B-deletion germline polymorphism in Taiwanese oral squamous cell carcinoma that impacts expression of APOBEC3A, and is shown to be of clinical prognostic relevance. Our finding might be recapitulated by genomic studies in other cancer types.Oral squamous cell carcinoma is a prevalent malignancy in Taiwan. Here, the authors show that OSCC in Taiwanese show a frequent deletion polymorphism in the cytidine deaminases gene cluster APOBEC3 resulting in increased expression of A3A, which is shown to be of clinical prognostic relevance.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Taiwan Cancer Registry. Health Promotion Administration, Ministry of Health and Welfare, Taiwan. Available: http://tcr.cph.ntu.edu.tw/main.php?Page=A1. Accessed 6 May 2015.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

