Multivariate discovery and replication of five novel loci associated with Immunoglobulin G N-glycosylation
- PMID: 28878392
- PMCID: PMC5587582
- DOI: 10.1038/s41467-017-00453-3
Multivariate discovery and replication of five novel loci associated with Immunoglobulin G N-glycosylation
Abstract
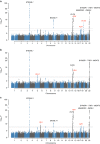
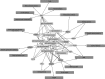
Joint modeling of a number of phenotypes using multivariate methods has often been neglected in genome-wide association studies and if used, replication has not been sought. Modern omics technologies allow characterization of functional phenomena using a large number of related phenotype measures, which can benefit from such joint analysis. Here, we report a multivariate genome-wide association studies of 23 immunoglobulin G (IgG) N-glycosylation phenotypes. In the discovery cohort, our multi-phenotype method uncovers ten genome-wide significant loci, of which five are novel (IGH, ELL2, HLA-B-C, AZI1, FUT6-FUT3). We convincingly replicate all novel loci via multivariate tests. We show that IgG N-glycosylation loci are strongly enriched for genes expressed in the immune system, in particular antibody-producing cells and B lymphocytes. We empirically demonstrate the efficacy of multivariate methods to discover novel, reproducible pleiotropic effects.Multivariate analysis methods can uncover the relationship between phenotypic measures characterised by modern omic techniques. Here the authors conduct a multivariate GWAS on IgG N-glycosylation phenotypes and identify 5 novel loci enriched in immune system genes.
Conflict of interest statement
The authors declare no competing financial interests. G.L. has multiple patents in the field of glycoscience issued. Y.S.A. is a director and co-owner of Maatschap PolyOmica, which provides (consulting) services in the area of (statistical) (gen)omics.
Figures


References
-
- Astle W, Balding DJ. Population structure and cryptic relatedness in genetic association studies. Stat. Sci. 2009;24:451–471. doi: 10.1214/09-STS307. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

