Differences in Growth Properties among Two Human Cytomegalovirus Glycoprotein O Genotypes
- PMID: 28878758
- PMCID: PMC5572245
- DOI: 10.3389/fmicb.2017.01609
Differences in Growth Properties among Two Human Cytomegalovirus Glycoprotein O Genotypes
Abstract
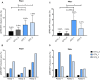
Glycoprotein O (gO) of the human cytomegalovirus (HCMV) is the critical subunit of the envelope trimer gH/gL/gO as it interacts with platelet-derived growth factor alpha receptor upon fibroblast entry, and triggers gB-mediated fusion for fibroblast and epithelial cell infection. Eight genotypes (GT) of the highly polymorphic gO gene are described, yet it is unclear whether the distinct GTs differ in their function. Thus, we aimed to elucidate potential functional differences between two highly diverse gO GTs in an otherwise genomically identical HCMV strain. Therefore, resident gO GT1c sequence of strain TB40-BAC4-luc was entirely replaced by gO GT4 of strain Towne and both, GT1c and GT4 viruses, were investigated for their growth properties in fibroblasts and epithelial cells. In addition, two conserved gO cysteines involved in gH/gL/gO stabilization were mutated to serine either in GT1c (C218S and C343S) or GT4 (C216S and C336S) and their effects on cell-free infectivity were assessed. GT4 viruses displayed a significantly enhanced epithelial cell tropism and this resulted in higher virus release upon replication in epithelial cells when compared to GT1c viruses. Further, when the two cysteines were individually mutated in gO GT1c no impairment in cell-free infectivity was observed. This, however, was in sharp contrast to gO GT4, in which both of the corresponding cysteine mutations led to a substantial reduction in cell-free infectivity which was even more pronounced upon mutation of GT4-C336 than of GT4-C216. In conclusion, these findings provide evidence that the two highly diverse gO genotypes, GT1c and GT4, differ in their functional properties as revealed by their different infection capacities for epithelial cells and by their different responsiveness to mutation of strictly conserved cysteine residues. Thus, it is likely that the gO heterogeneity influences cell-free infectivity of HCMV also in vivo which may have important implications for virus host transmission.
Keywords: HCMV; epithelial cells; fibroblasts; glycoprotein O; trimeric complex; tropism.
Figures





References
-
- Chen J. Y., Zheng T. L., Zhou T., Hu P. W., Huang M. J., Xu X., et al. (2016). Human cytomegalovirus prevalence and distribution of glycoprotein B, O genotypes among hospitalized children with respiratory infections in West China, 2009-2014. Trop. Med. Int. Health 21 1428–1434. 10.1111/tmi.12770 - DOI - PubMed
-
- Ciferri C., Chandramouli S., Donnarumma D., Nikitin P. A., Cianfrocco M. A., Gerrein R., et al. (2015). Structural and biochemical studies of HCMV gH/gL/gO and Pentamer reveal mutually exclusive cell entry complexes. Proc. Natl. Acad. Sci. U.S.A. 112 1767–1772. 10.1073/pnas.1424818112 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

