Exploiting sequence-based features for predicting enhancer-promoter interactions
- PMID: 28881991
- PMCID: PMC5870728
- DOI: 10.1093/bioinformatics/btx257
Exploiting sequence-based features for predicting enhancer-promoter interactions
Abstract
Motivation: A large number of distal enhancers and proximal promoters form enhancer-promoter interactions to regulate target genes in the human genome. Although recent high-throughput genome-wide mapping approaches have allowed us to more comprehensively recognize potential enhancer-promoter interactions, it is still largely unknown whether sequence-based features alone are sufficient to predict such interactions.
Results: Here, we develop a new computational method (named PEP) to predict enhancer-promoter interactions based on sequence-based features only, when the locations of putative enhancers and promoters in a particular cell type are given. The two modules in PEP (PEP-Motif and PEP-Word) use different but complementary feature extraction strategies to exploit sequence-based information. The results across six different cell types demonstrate that our method is effective in predicting enhancer-promoter interactions as compared to the state-of-the-art methods that use functional genomic signals. Our work demonstrates that sequence-based features alone can reliably predict enhancer-promoter interactions genome-wide, which could potentially facilitate the discovery of important sequence determinants for long-range gene regulation.
Availability and implementation: The source code of PEP is available at: https://github.com/ma-compbio/PEP .
Contact: jianma@cs.cmu.edu.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2017. Published by Oxford University Press. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com
Figures
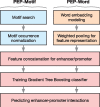
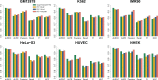

References
-
- Bonev B., Cavalli G. (2016) Organization and function of the 3d genome. Nat. Rev. Genet., 17, 661–678. - PubMed
-
- Chen T., Guestrin C. (2016a) XGBoost. https://github.com/dmlc/xgboost.
-
- Chen T., Guestrin C. (2016b) XGBoost: A scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, p.785–794. ACM, New York, NY, USA.
-
- Davis J., Goadrich M. (2006) The relationship between precision-recall and ROC curves. In: Proceedings of the 23rd International Conference on Machine Learning, p.233–240. ACM, New York, NY, USA.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

