Apico-basal Polarity Determinants Encoded by crumbs Genes Affect Ciliary Shaft Protein Composition, IFT Movement Dynamics, and Cilia Length
- PMID: 28882989
- PMCID: PMC5676222
- DOI: 10.1534/genetics.117.300260
Apico-basal Polarity Determinants Encoded by crumbs Genes Affect Ciliary Shaft Protein Composition, IFT Movement Dynamics, and Cilia Length
Erratum in
-
Apico-basal Polarity Determinants Encoded by crumbs Genes Affect Ciliary Shaft Protein Composition, IFT Movement Dynamics, and Cilia Length.Genetics. 2019 Aug;212(4):1469. doi: 10.1534/genetics.119.302460. Genetics. 2019. PMID: 31405998 Free PMC article. No abstract available.
Abstract
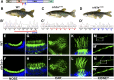
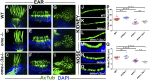
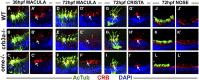
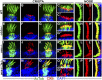
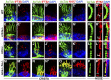
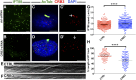
One of the most obvious manifestations of polarity in epithelia is the subdivision of the cell surface by cell junctions into apical and basolateral domains. crumbs genes are among key regulators of this form of polarity. Loss of crumbs function disrupts the apical cell junction belt and crumbs overexpression expands the apical membrane size. Crumbs proteins contain a single transmembrane domain and localize to cell junction area at the apical surface of epithelia. In some tissues, they are also found in cilia. To test their role in ciliogenesis, we investigated mutant phenotypes of zebrafish crumbs genes. In zebrafish, mutations of three crumbs genes, oko meduzy/crb2a, crb3a, and crb2b, affect cilia length in a subset of tissues. In oko meduzy (ome), this is accompanied by accumulation of other Crumbs proteins in the ciliary compartment. Moreover, intraflagellar transport (IFT) particle components accumulate in the ciliary shaft of ome;crb3a double mutants. Consistent with the above, Crb3 knockdown in mammalian cells affects the dynamics of IFT particle movement. These findings reveal crumbs-dependent mechanisms that regulate the localization of ciliary proteins, including Crumbs proteins themselves, and show that crumbs genes modulate intraflagellar transport and cilia elongation.
Keywords: Crumbs; IFT; apico-basal polarity; cilia; cristae.
Copyright © 2017 by the Genetics Society of America.
Figures

References
-
- Chih B., Liu P., Chinn Y., Chalouni C., Komuves L. G., et al. , 2011. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat. Cell Biol. 14: 61–72. - PubMed
-
- den Hollander A. I., ten Brink J. B., de Kok Y. J., van Soest S., van den Born L. I., et al. , 1999. Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12). Nat. Genet. 23: 217–221. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

