Phenotype in combination with genotype improves outcome prediction in acute myeloid leukemia: a report from Children's Oncology Group protocol AAML0531
- PMID: 28883080
- PMCID: PMC5709105
- DOI: 10.3324/haematol.2017.169029
Phenotype in combination with genotype improves outcome prediction in acute myeloid leukemia: a report from Children's Oncology Group protocol AAML0531
Abstract
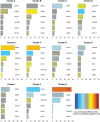
Diagnostic biomarkers can be used to determine relapse risk in acute myeloid leukemia, and certain genetic aberrancies have prognostic relevance. A diagnostic immunophenotypic expression profile, which quantifies the amounts of distinct gene products, not just their presence or absence, was established in order to improve outcome prediction for patients with acute myeloid leukemia. The immunophenotypic expression profile, which defines each patient's leukemia as a location in 15-dimensional space, was generated for 769 patients enrolled in the Children's Oncology Group AAML0531 protocol. Unsupervised hierarchical clustering grouped patients with similar immunophenotypic expression profiles into eleven patient cohorts, demonstrating high associations among phenotype, genotype, morphology, and outcome. Of 95 patients with inv(16), 79% segregated in Cluster A. Of 109 patients with t(8;21), 92% segregated in Clusters A and B. Of 152 patients with 11q23 alterations, 78% segregated in Clusters D, E, F, G, or H. For both inv(16) and 11q23 abnormalities, differential phenotypic expression identified patient groups with different survival characteristics (P<0.05). Clinical outcome analysis revealed that Cluster B (predominantly t(8;21)) was associated with favorable outcome (P<0.001) and Clusters E, G, H, and K were associated with adverse outcomes (P<0.05). Multivariable regression analysis revealed that Clusters E, G, H, and K were independently associated with worse survival (P range <0.001 to 0.008). The Children's Oncology Group AAML0531 trial: clinicaltrials.gov Identifier: 00372593.
Trial registration: ClinicalTrials.gov NCT00372593.
Copyright© 2017 Ferrata Storti Foundation.
Figures




References
-
- Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. - PubMed
-
- Clinical and Laboratory Standards Institute. Clinical flow cytometric analysis of neoplas tic hematolymphoid cells; approved guideline—Second edition. Wayne, PA: US Food and Drug Administration; 2008. FDA publication no. 7-150.
-
- Loken MR. Residual disease in AML, a target that can move in more than one direction [editorial]. Cytometry B. 2014; 86(1):15–17. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

