A thiol probe for measuring unfolded protein load and proteostasis in cells
- PMID: 28883394
- PMCID: PMC5589734
- DOI: 10.1038/s41467-017-00203-5
A thiol probe for measuring unfolded protein load and proteostasis in cells
Abstract
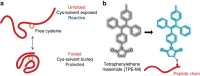
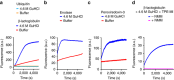
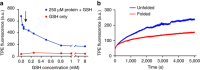
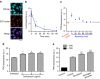
When proteostasis becomes unbalanced, unfolded proteins can accumulate and aggregate. Here we report that the dye, tetraphenylethene maleimide (TPE-MI) can be used to measure cellular unfolded protein load. TPE-MI fluorescence is activated upon labelling free cysteine thiols, normally buried in the core of globular proteins that are exposed upon unfolding. Crucially TPE-MI does not become fluorescent when conjugated to soluble glutathione. We find that TPE-MI fluorescence is enhanced upon reaction with cellular proteomes under conditions promoting accumulation of unfolded proteins. TPE-MI reactivity can be used to track which proteins expose more cysteine residues under stress through proteomic analysis. We show that TPE-MI can report imbalances in proteostasis in induced pluripotent stem cell models of Huntington disease, as well as cells transfected with mutant Huntington exon 1 before the formation of visible aggregates. TPE-MI also detects protein damage following dihydroartemisinin treatment of the malaria parasites Plasmodium falciparum. TPE-MI therefore holds promise as a tool to probe proteostasis mechanisms in disease.Proteostasis is maintained through a number of molecular mechanisms, some of which function to protect the folded state of proteins. Here the authors demonstrate the use of TPE-MI in a fluorigenic dye assay for the quantitation of unfolded proteins that can be used to assess proteostasis on a cellular or proteome scale.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
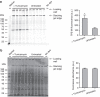

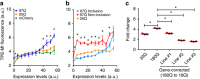
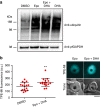
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

